Embedding adapters, anchor molecules, molecular membranes, devices, methods and applications
A molecular membrane and molecular technology, applied in chemical instruments and methods, biochemical equipment and methods, and microbial determination/inspection, etc., can solve problems such as affecting the detection effect, the length of effective current data output, and the difficulty of detecting biomolecules. To achieve the effect of increasing the output time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Example 1: Preparation of an anchor molecule
[0100] Design and synthesize linker, its nucleotide sequence is as follows:
[0101] GGTCGGTGCTGGACTTTTTTTTTTTTTTTTTTTTT-DBCO;
[0102] Design and synthesize the embedded linker, its amino acid sequence is as follows:
[0103] N3-WERNLP- SVSGLLKIIGFSTSVTALGFVLY -KYKLL PRS.
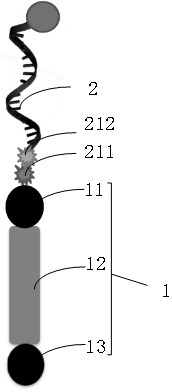
[0104] In this embodiment, the connection linker is connected to the analyte through the capture chain, the insertion linker is a polypeptide, and the connection linker and the insertion linker are connected through "DBCO" and "N3". see Figure 4 , is the predicted map of the polypeptide transmembrane region, from Figure 4 It can be seen that the 7th to 29th amino acids in the amino acid sequence (underlined in the amino acid sequence) are transmembrane regions, indicating that the anchor molecule can penetrate the amphiphile membrane.
[0105] The connecting adapter is incubated with the embedding adapter, and the incubated product is collected...
Embodiment 2
[0106] Example 2: Preparation and detection of an embedded joint
[0107] Design and synthesize the embedded linker, its amino acid sequence is as follows:
[0108] DBCO-YIK- LFIMIVGGLVGLRIVFAVLSIVN -RVRQ.
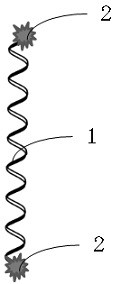
[0109] see Figure 7 , is the prediction map of the transmembrane region of the polypeptide, wherein amino acids 4 to 26 (underlined in the amino acid sequence) are transmembrane regions.
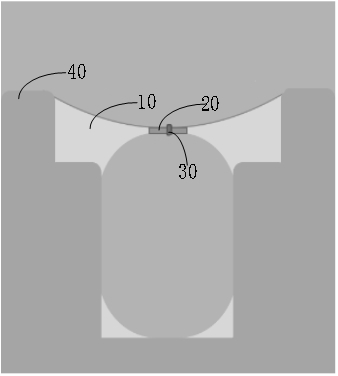
[0110] Connect the fluorophore FAM or Cy5 to the embedding adapter, add a buffer solution (500 mM KCl, 20 mM HEPES, pH 8.0) into the reservoir chamber of the biochip of the gene sequencer QNome-9604 of Qitan Technology Co., Ltd., and then Add 40 μl of silicone oil and phosphatidylcholine (PC, 10mg / mL) mixed oil, and finally add the aforementioned buffer solution to prepare a molecular film, the structure of which is as follows: figure 2 Shown; Mix the embedded linker with the fluorescent group and the molecular membrane, so that the concentration of the embedded linker in the solu...
Embodiment 3
[0112] Example 3: Nanopore sequencing of analytes using anchor molecules
[0113] (1) Set up an experimental group and a control group. In the experimental group and the control group, the same biochip was used, and the molecular film was prepared according to the method in Example 2;
[0114] (2) On the molecular membrane of the experimental group, add the anchor molecule prepared in Example 1, so that the concentration of the embedded linker in the solution on the adding side of the molecular membrane is 0.5 μM, and incubate at room temperature for 60 minutes; on the molecular membrane of the control group , without adding any anchor molecules, incubate at room temperature for 60 minutes;
[0115] (3) Samples of the same concentration were added to both the experimental group and the control group, and the gene sequencer QNome-9604 of Qitan Technology Co., Ltd. was used to sequence the ssDNA in the samples to obtain the via signal map.
[0116] see Figure 10 , Figure 11...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com