Method for obtaining driving element
A technology of driving elements and promoters, applied in the field of genetic engineering, can solve the problems of inability to distinguish cancer cell subsets, systemic recurrence of patients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0075] The preparation process in step 3 and step 4, such as the means of knockout, insertion and functional connection at specific sites, such as CRISPR technology, CRISPR-Cas9 technology, etc., are well known to those skilled in the art and will not be repeated here.
[0076] In step 5, when the selected cancer cell line is a human highly metastatic liver cancer cell line with the registration number MHCC997H, the specific implementation method can be to uniformly mix different cell phenotype state evaluators in the form of lentiviral vectors and then infect them to be studied For the target cells, use Blasticidin to screen out stable liver cancer cell lines. Of course, when the cancer cell lines used are other cancer cell lines, the corresponding screening methods should be adjusted accordingly, which will not be repeated here.
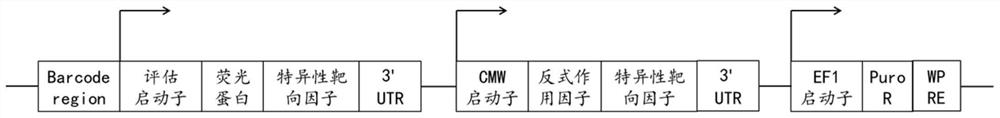
[0077] In step 6, after the constructed synthetic promoter lentiviral test vector is used to infect the stable cancer cell line containing the cel...
Embodiment
[0108] Material
[0109] cell:
[0110] MHCC97H human highly metastatic liver cancer cell line was provided by Shanghai Institute of Cell Biology, Chinese Academy of Sciences.
[0111] Drugs and reagents:
[0112] Doxorubicin (Adriamycin), specification: 10mg, batch number: KFS276, supplier: Biolab.
[0113] Sorafenib (Sorafenib) specification: 10mg, batch number: Bay 43-9006, supplier: Shanghai Ruihui Chemical Technology Co., Ltd.
[0114] Roseville-1640 (RPMI-1640) medium and fetal bovine serum (FBS) were purchased from GIBCO, USA.
[0115] Doxycycline (doxycycline hydrochloride), specification: 10mM / mL, batch number: ID0390-10mM*1mL (inWater), supplier: Suo Laibao.
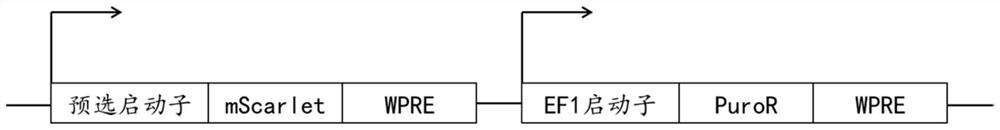
[0116] Plasmids were synthesized from General Biology (Anhui), the classifier plasmid backbone used SWB-Blasticidin lentiviral backbone, and the synthetic promoter vector backbone used SWP-Puromycin.
[0117] Lipofectamine 3000, specification: 1mL, batch number: L3000015, supplier: ThermalFisher.
[0118]...
Embodiment approach
[0122] MHCC97H liver cancer cells were cultured in RPMI-1640 containing 10% FBS, 0.1 μM Doxorubicin and 5 μM Sora fenib at 37°C, 5% CO 2 cultured in an incubator. Passage once every 3 days, and take cells in the logarithmic growth phase to perform high-throughput functional screening of drug-resistant synthetic promoters to obtain multiple drug-resistant cancer cell subpopulations.
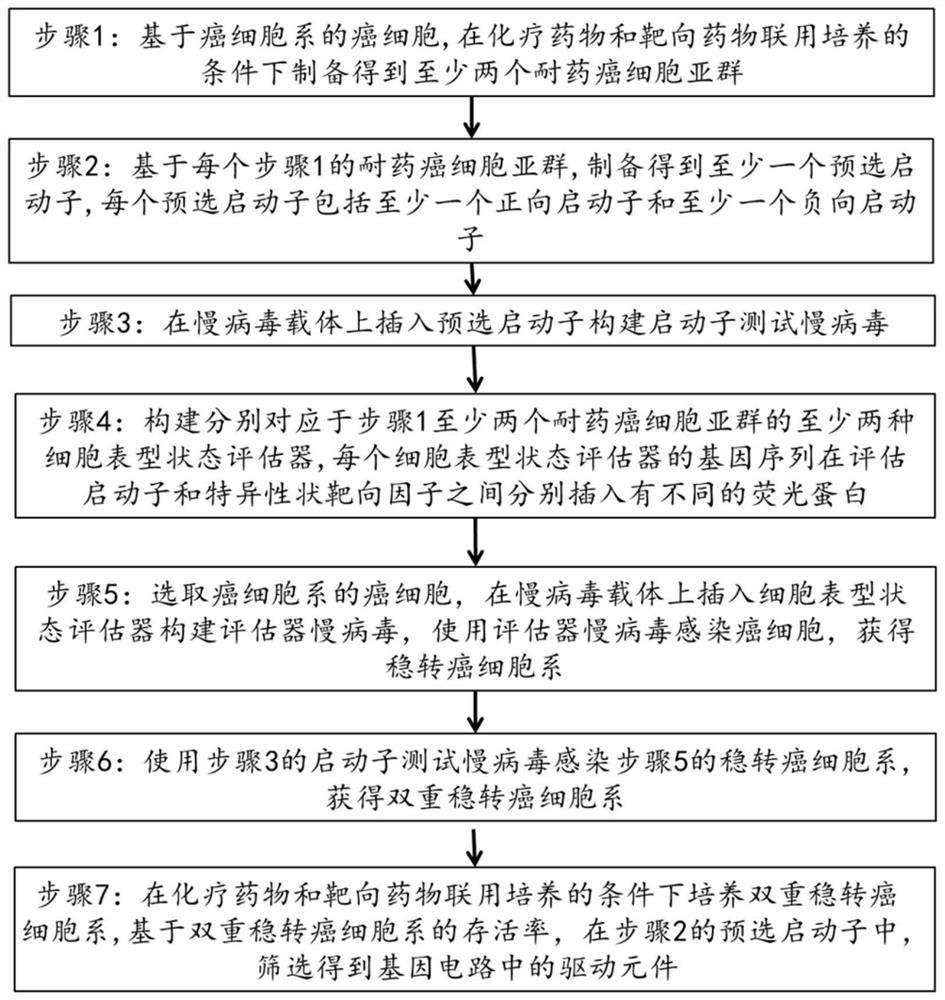
[0123] Screen the candidate promoters with positive (activity from none) or negative (activity to no) activity indication reactions from the obtained drug-resistant cancer cell subpopulations, and label the screened candidate promoters sequentially For sample 1 to sample 10 packs containing such as figure 2 Promoter test lentiviruses for the indicated gene sequences.
[0124] After successful packaging of the lentivirus, the lentiviral supernatant was collected and filtered through a 0.45 μm filter to remove cells and debris.
[0125] Mix lentiviral supernatant (4 parts) and 5X lentiviral conc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com