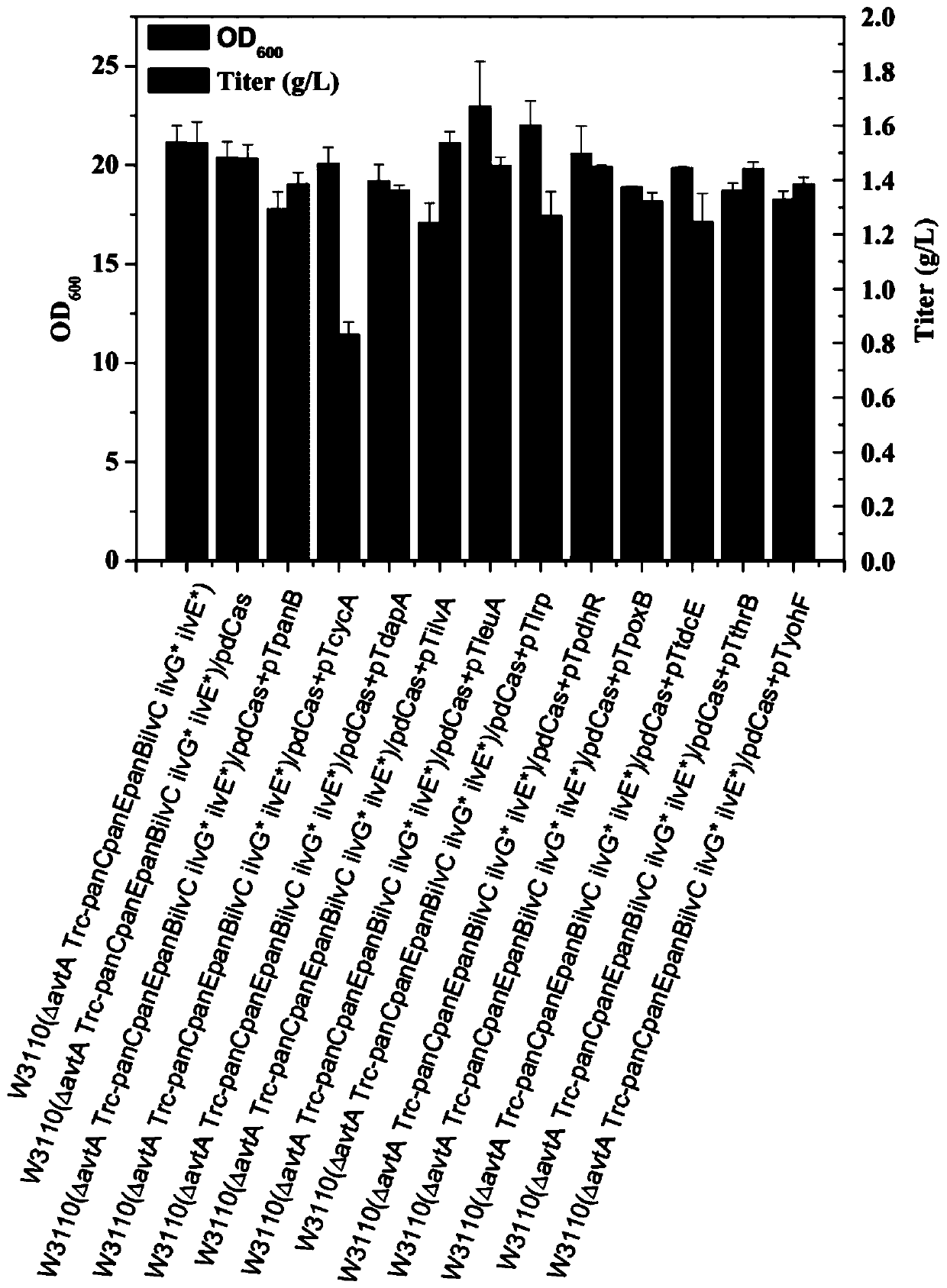
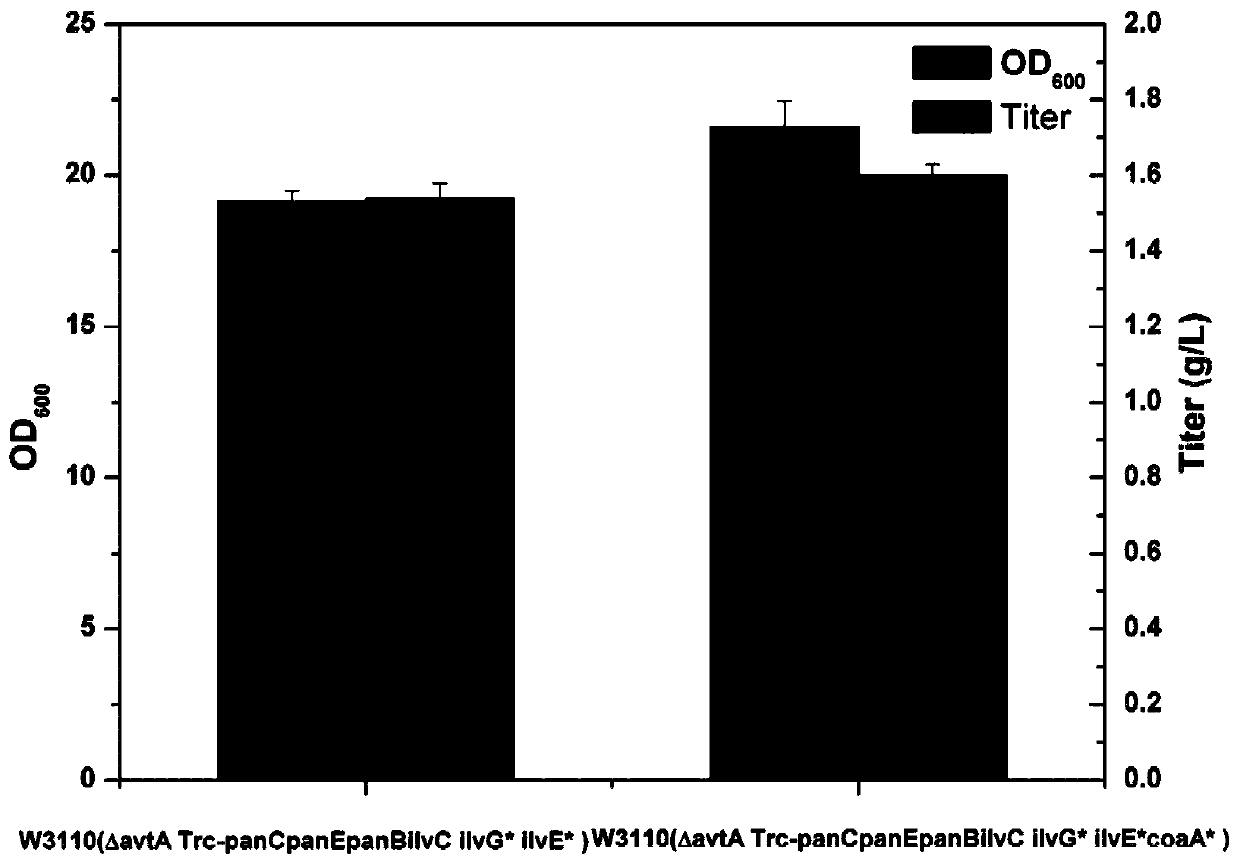
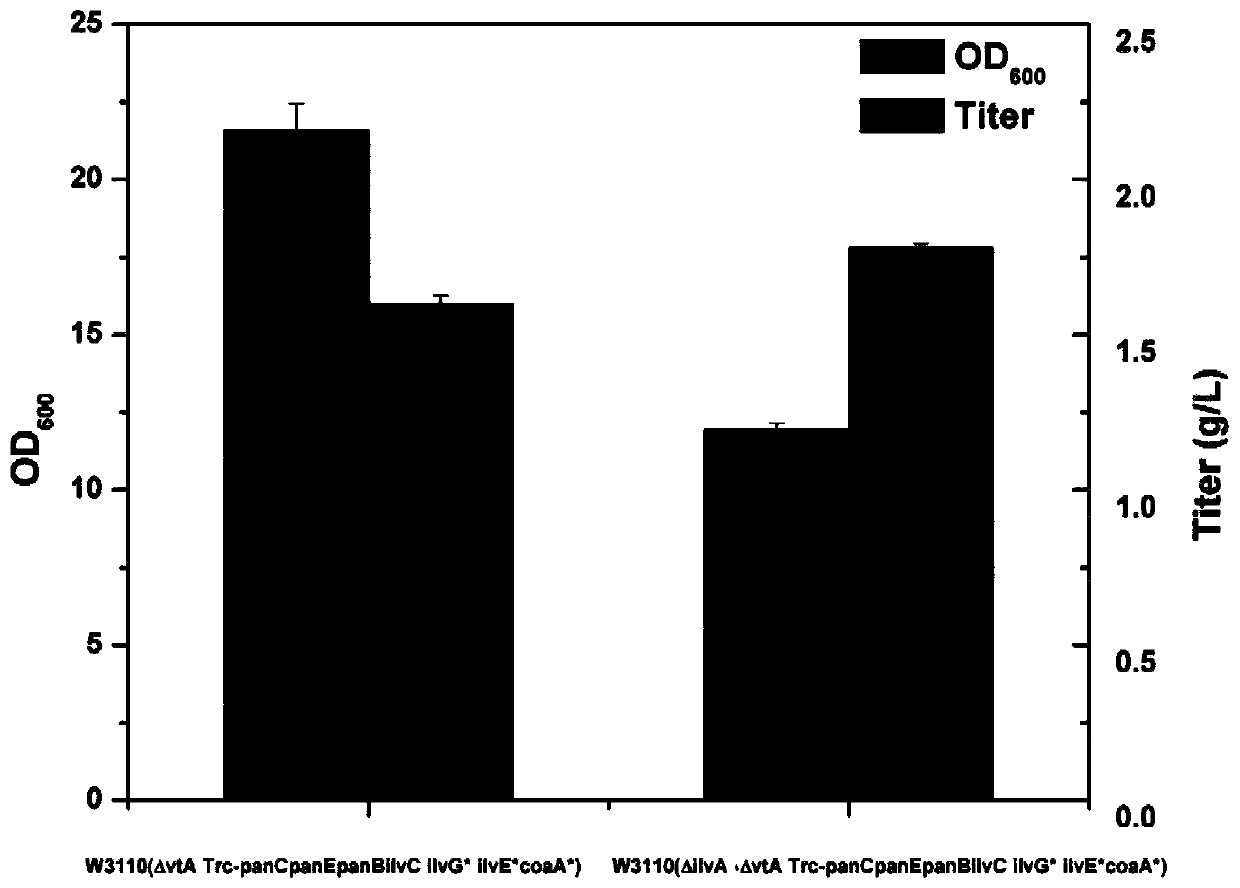
Genetic engineering bacteria capable of producing pantothenic acid at high yield without addition of beta-alanine, construction and application of genetic engineering bacteria
A technology of genetically engineered bacteria and alanine, applied in the direction of genetic engineering, application, plant genetic improvement, etc., can solve the problem that the research on D-pantothenic acid fermentation is not in-depth, and achieve the effect of enhancing the utilization capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0062] Embodiment 1: the HPLC determination of D-pantothenic acid content
[0063] The detection method is as follows:
[0064] Chromatographic conditions: C 18 Column (250×4.6mm, particle size 5μm, Agilent Technologies Co., Santa Clara, CA, USA), detection wavelength: 200nm, column temperature: 30°C;
[0065] Sample treatment: Dilute the sample with ultrapure water to keep the D-pantothenic acid content between 0.05g / L and 0.40g / L;
[0066] Mobile phase: acetonitrile / water / phosphoric acid: (50 / 949 / 1);
[0067] Data collection time: 18min.
Embodiment 2
[0068] Example 2: Construction and shake flask fermentation of effectively utilizing extracellular β-alanine strain W3110 (Trc-panC)
[0069] Using Escherichia coli W3110 as the starting strain, the gene editing technology mediated by CRISPR-Cas9 was used (Yu Jiang et al.2015Multigene Editing in the Escherichia coli Genome via the CRISPR-Cas9System.Applied Environmental Microbiology.81:2506-2514), derived from The trc promoter of pTrc99A (nucleotide sequence shown in SEQ ID No.1) replaces the original promoter of panC (GeneID: 12932172) in the genome to enhance the expression intensity of panC.
[0070] (1) Construction of pTarget-panC plasmid: Use pTarget F plasmid (Addgene Plasmid#62226) as a template, use pTarget-panC-1 / pTarget-panC-2 as primers for PCR amplification, and the PCR product is digested by Dpn I at 37°C 3h, then transformed into E.coli DH5α, screened with spectacle enzyme plate, and sequenced to verify that the correct pTarget-panC plasmid was obtained, which w...
Embodiment 3
[0078] Example 3: Construction of high-expression ketopantoate reductase strain W3110 (Trc-panCpanE) and shake flask fermentation
[0079] (1) Construction of pTarget-panE plasmid: Use pTarget F plasmid (Addgene Plasmid#62226) as a template, use pTarget-panE-1 / pTarget-panE-2 as primers for PCR amplification, and digest the PCR product with Dpn I at 37°C 3h, then transformed into E.coli DH5α, screened with spectacle enzyme plate, and sequenced to verify that the correct pTarget-panE plasmid was obtained, which was used for subsequent connection of DonorDNA.
[0080] (2) Construction of pTD-panE plasmid: with the E.coli W3110 genome as a template, pTD-panE-1, pTD-panE-2, pTD-panE-3 and pTD-panE-4 as primers, the construction steps are the same as in Example 2 (2), obtain pTD-panE plasmid.
[0081] (3) The pCas plasmid (Addgene Plasmid #62225) was introduced into the competent W3110 (Trc-panC) obtained in Example 2, and the preparation method of the competent W3110 (Trc-panC) wa...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com