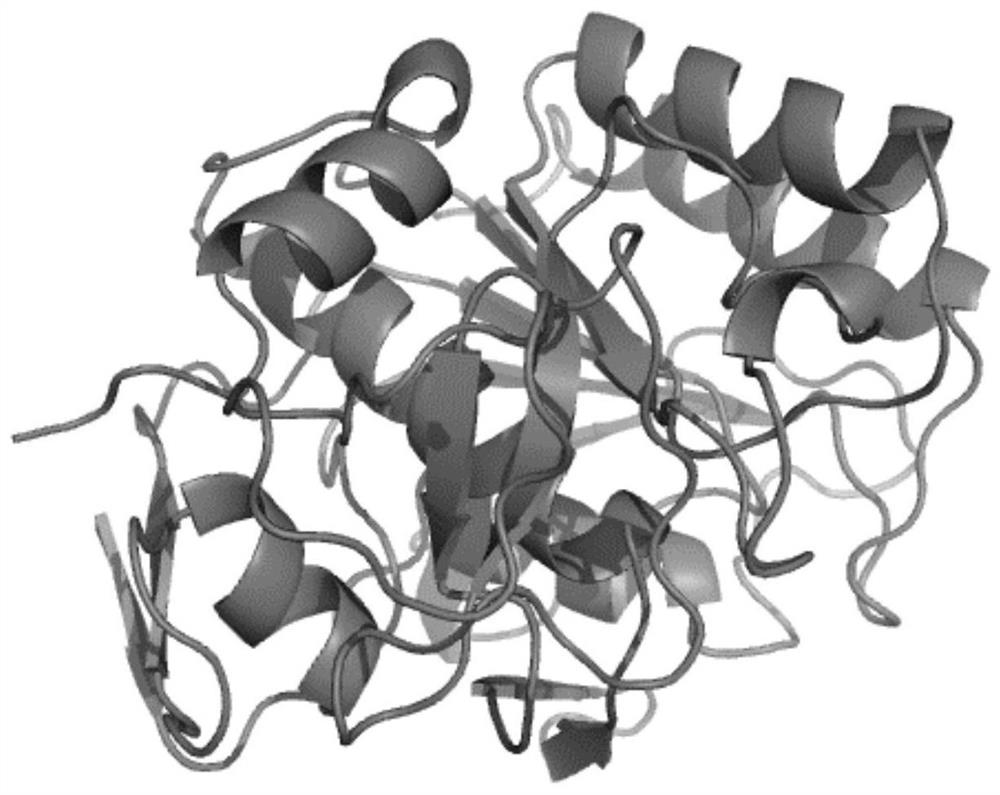
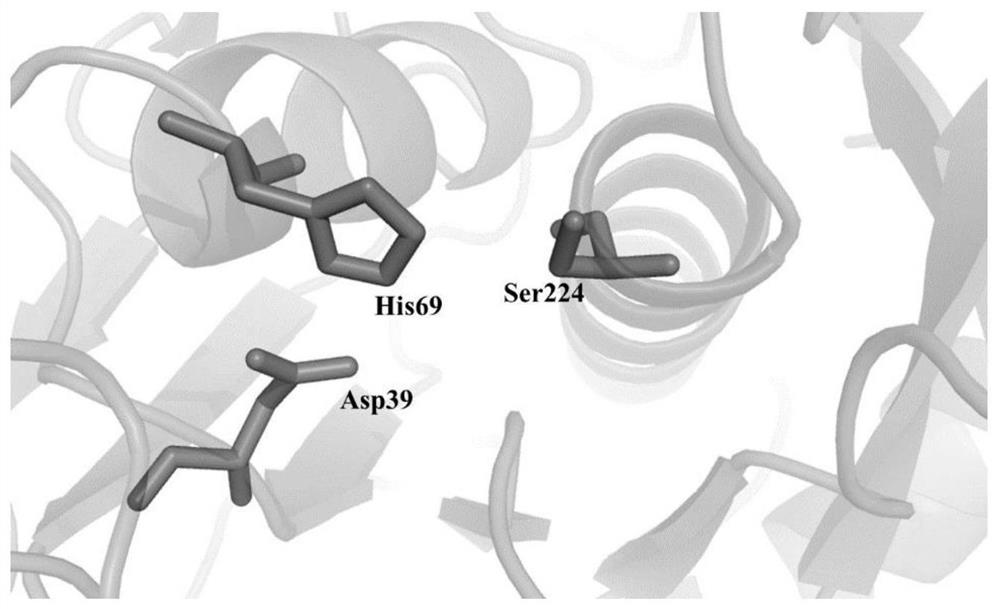
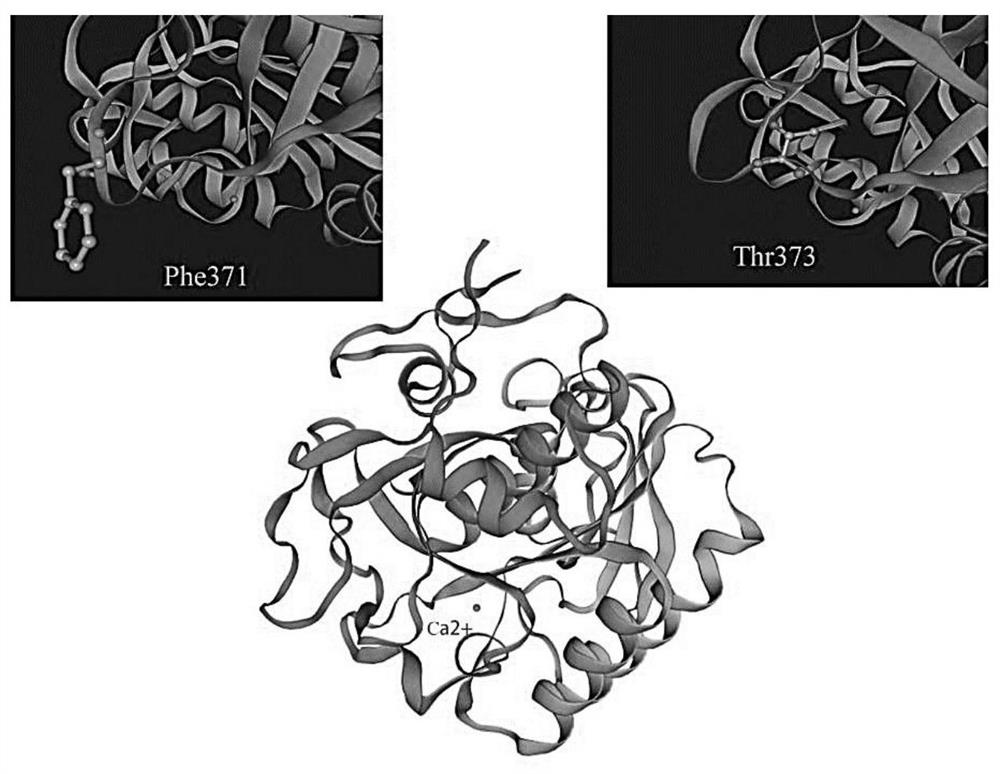
Protease K mutant and construction and application of expression vector thereof
A protease and mutant technology, which is applied in the field of construction and application of proteinase K mutants and their expression vectors, can solve the problems of slow production of Candida albicans limberii, limitation of actual production of proteinase K, unfavorable industrial production, etc., and achieve improvement Screening for effects of weighing power, reduced handling, good enzyme activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0043] The embodiment of the present invention provides the preparation method of the proteinase K mutant as described in any of the foregoing embodiments, which includes: cultivating the genetically engineered bacteria as described in the foregoing embodiments or the gene constructed by the construction method described in any of the foregoing embodiments Engineering bacteria to induce the expression of the proteinase K mutant; or, on the basis of the amino acid sequence shown in SEQ ID No.1, carry out at least one of the following mutations: F371C and T373P.
[0044] In addition, the embodiments of the present invention also provide proteinase K mutants as described in any of the foregoing embodiments or genetically engineered bacteria as described in the foregoing embodiments, or genetically engineered bacteria constructed by the construction method described in any of the foregoing embodiments can be hydrolyzed Application in protein or preparation of products for hydrolysi...
Embodiment 1
[0047] A proteinase K mutant, its amino acid sequence is shown in SEQ ID No.2, and its nucleic acid sequence is shown in SEQ ID No.3.
[0048] The preparation method of the above proteinase K mutant comprises the following steps.
[0049] 1. Construction of expression vector.
[0050] 1.1 Construction of single copy expression vector.
[0051] Using pPIC9k as the basic framework, the corresponding primers were designed according to the Golden gate assembly principle, as shown in Table 1. Six framework fragments were obtained by cloning with P202-2 high-fidelity polymerase from Nanjing Master Biotechnology Co., Ltd., which were named Frag1, Frag2, and Frag3. , Frag4, Frag5 and Frag6.
[0052] Table 1 Primer Sequence
[0053]
[0054]
[0055] Note: F is the upstream primer, R is the downstream primer.
[0056] The DNA fragments with high purity were recovered from the fragment gels obtained by the above clones, and Frag1 was fused with Frag2, Frag3 was fused with Frag...
Embodiment 2
[0076]A proteinase K mutant and its preparation method are roughly the same as in Example 1, the difference being that the strain screening steps are different. In this example, the strain screening is as follows: linearize the pATG9-3K plasmid vector, and transfer to In Pichia pastoris GS115 competent cells, high-copy high-expression strains were obtained by screening BMMY plates containing different concentrations of G418 and containing 1% casein, which specifically included the following steps:
[0077] The pATG9-3K plasmid vector was digested and linearized by SacI, and the target fragment was excised by 0.8% agarose nucleic acid electrophoresis, and the gel was recovered and purified;
[0078] The above-mentioned purified gene fragments were electrotransformed into Pichia GS115 competent cells, spread on MD plates and cultured at 28°C for 2-3 days, and the Pichia transformants on MD plates were transferred to cells containing different Concentration (50μg / ul~500μg / ul) G41...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com