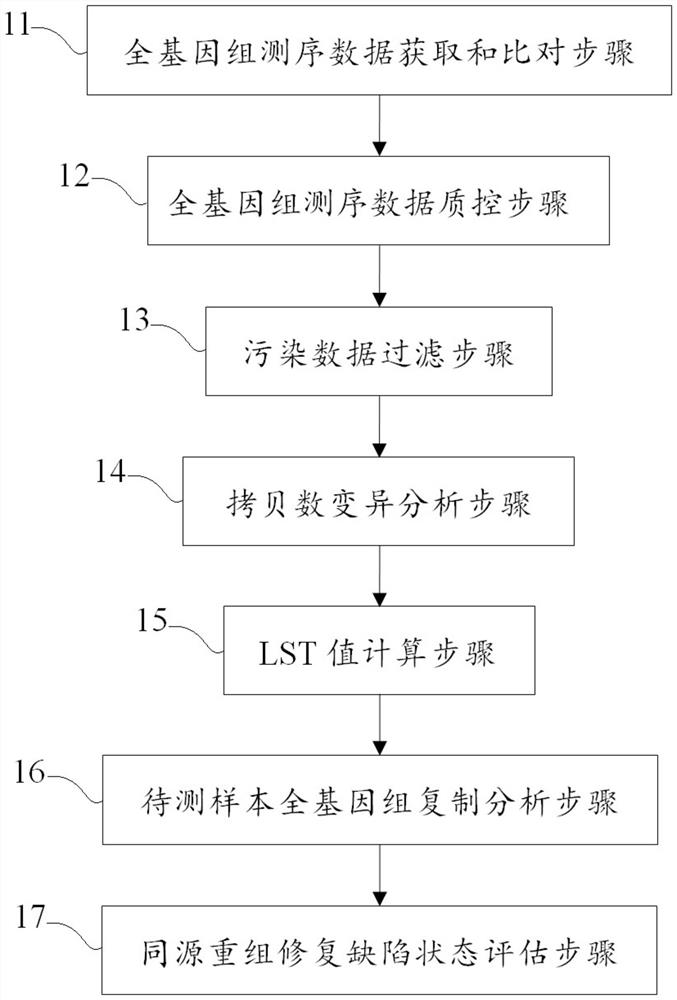
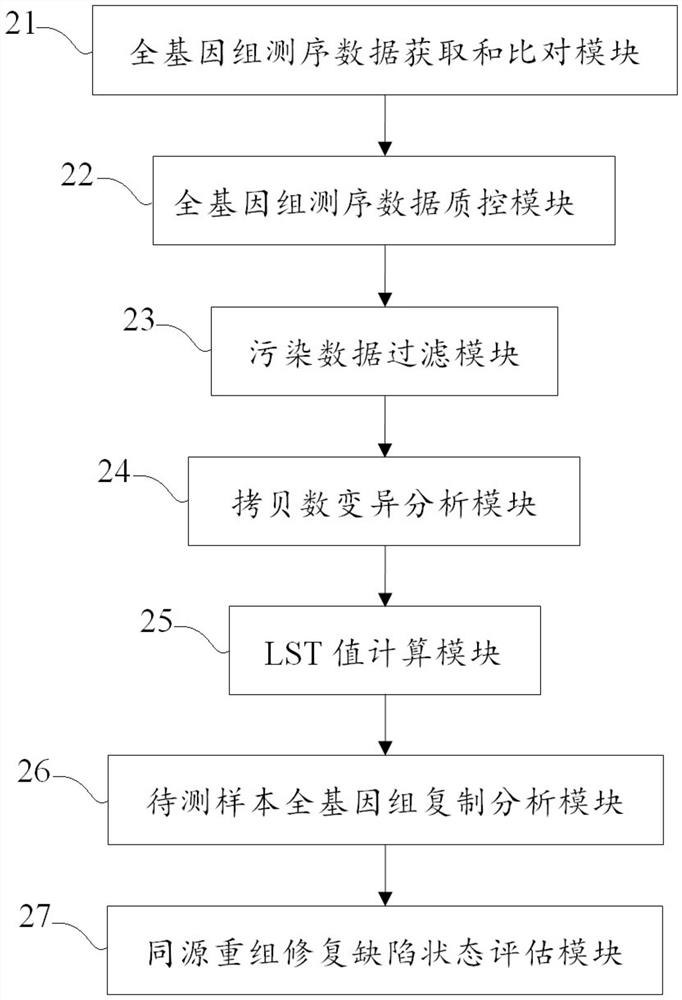
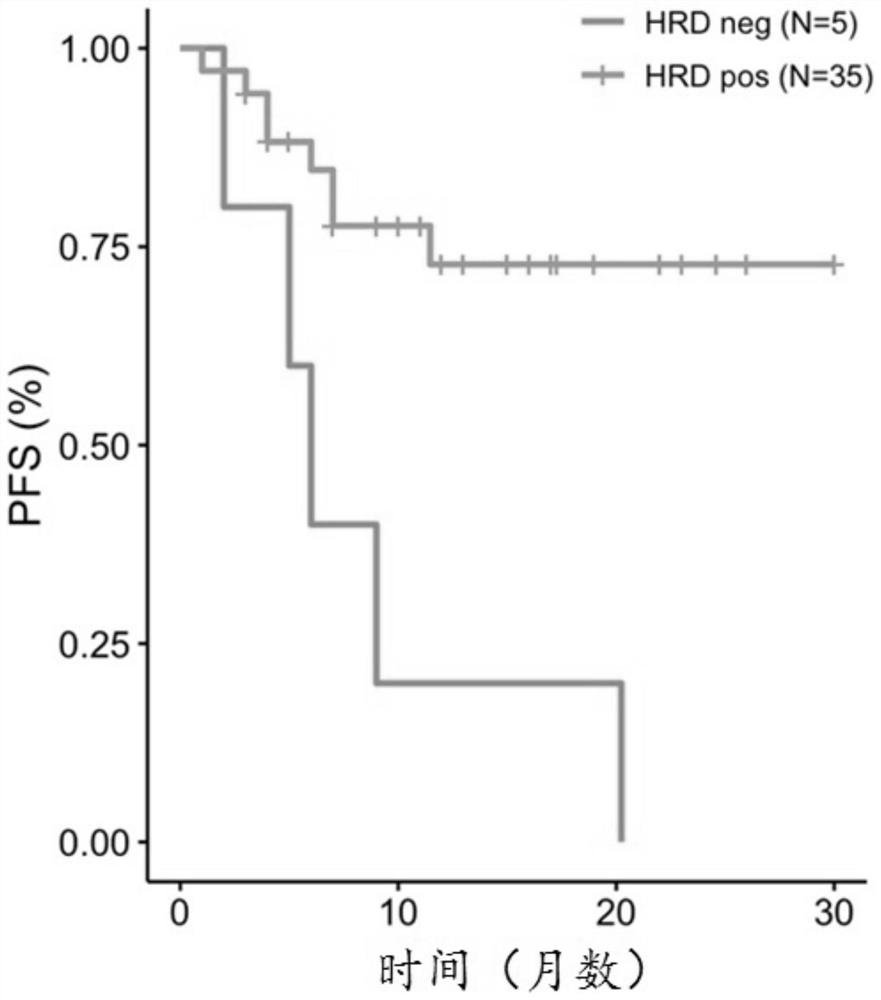
Evaluation method and device for homologous recombination repair defect and storage medium
A technology of homologous recombination and defects, applied in genomics, proteomics, instruments, etc., can solve problems such as poor performance, low sensitivity, and large errors in gene chip capture and sequencing data, so as to improve effectiveness and accuracy, calculate accurate effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0088] The tumor samples involved in this example were provided by Beijing Jiyinga Medical Laboratory Co., Ltd. A total of 40 real clinical samples with PARP efficacy, including 20 cases of BRCA wild type and 20 cases of BRCA mutant type. Methods for assessing homologous recombination repair deficiency in this example include:
[0089] (1) Nucleic acid extraction
[0090] In this example, tissue samples were used to extract DNA from 40 samples, TA cloning was used to connect adapters to build a library, and then Gene+Seq2000 was used for whole genome sequencing, with a data volume of 5G. get the raw data.
[0091] The silica gel membrane method was used for extraction, and DNA was extracted from formalin-fixed-paraffin-embedded (FFPE) tissues using the Generead DNA FFPE kitMinElute nucleic acid extraction column.
[0092] After extraction, the DNA was interrupted with a Diagenode Bioruptor Pico interrupter, so that the DNA was evenly interrupted to 200-250bp.
[0093] The ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com