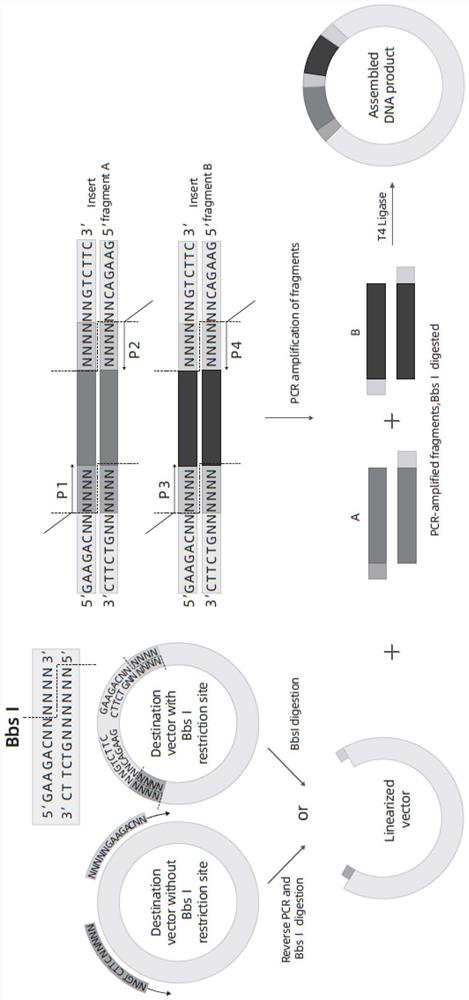
One-step method BbsI enzyme digestion connection fragment assembly method, assembly kit and application
An assembly method and kit technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of high time and labor costs, and achieve high assembly efficiency, high success rate, and optimized steps Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
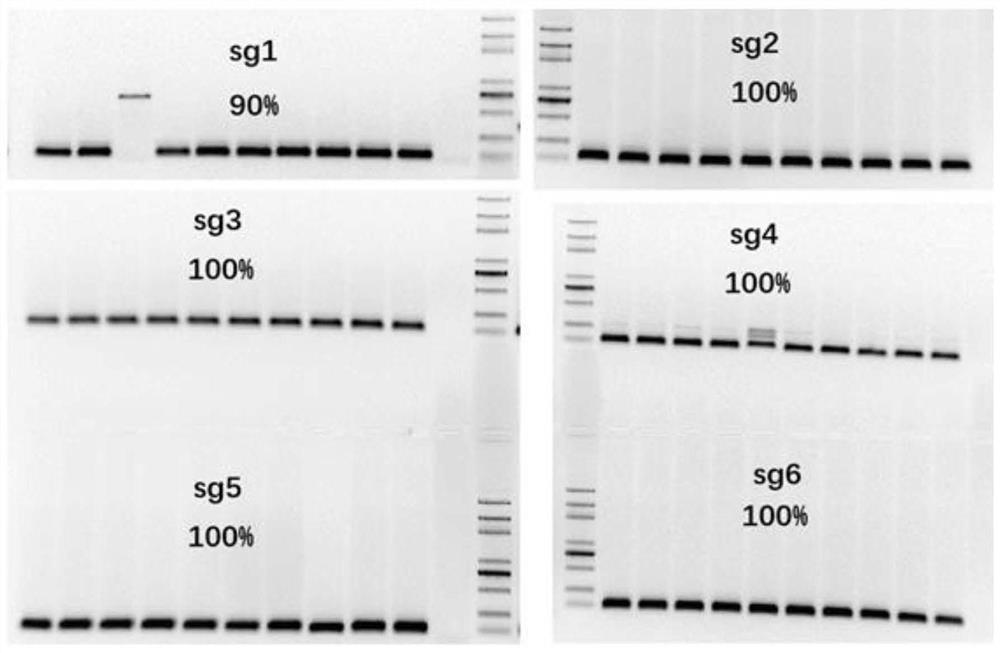
[0038] Example 1. Restriction ligation one-step method for encoding sgRNA sequence assembly
[0039] For the sequences of sgRNAs encoding targets of different species, use the one-step enzyme digestion ligation system to assemble the fragments onto the model plasmid, and explore the success probability of the ligation by colony PCR and sequencing. The specific experimental procedure is as follows.
[0040] 1. One pair of random targeting sequences, 2 pairs of mouse Wnt1 targeting sequences designed according to the website, and 3 pairs of targeting plant gRT, OsU6aT, and OsU3T sites in the references. The sequence information is shown in Table 1:
[0041] Table 1. Primers for sgRNA coding sequences of different targets
[0042]
[0043] 2. Configure the annealing system according to Table 2.
[0044] Table 2. Annealing reaction system
[0045]
[0046] Annealing reactions were performed according to Table 3.
[0047] Table 3. Annealing reaction program
[0048]
...
Embodiment 2
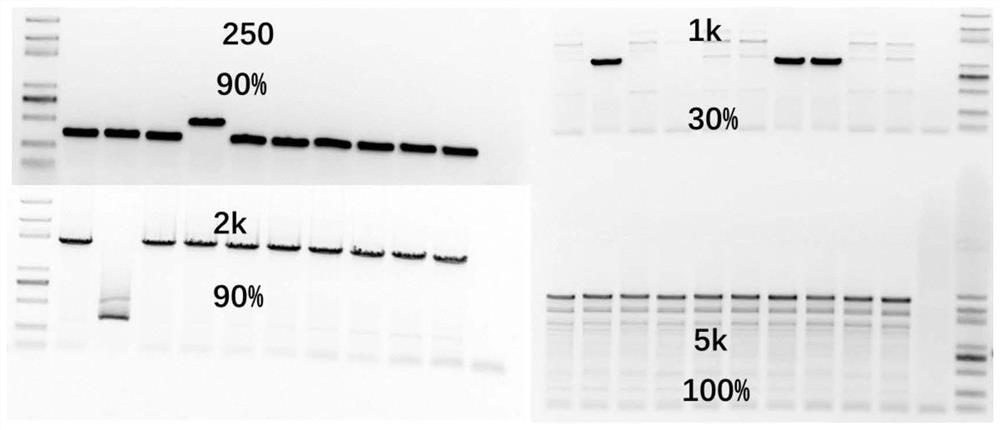
[0074] Example 2 The one-step method of enzyme digestion and ligation is used for the assembly of single fragments of different lengths
[0075] For the insertion fragments of different lengths, use the one-step enzyme digestion ligation system to assemble the fragments into approximately model plasmids, and explore the probability of successful connection by colony PCR and sequencing. The specific experimental procedure is as follows.
[0076] 1. Design insert fragments of different lengths, and the sequence information is shown in Table 7.
[0077] Table 7. Primer sequences for amplification of fragments of different lengths
[0078]
[0079] 2. Configure the amplification system according to Table 8.
[0080] Table 8. Amplification system
[0081]
[0082] Set up the amplification program of the model vector according to Table 9.
[0083] Table 9. Amplification Procedure
[0084]
[0085] 3. Purification of amplified fragments
[0086] Prepare a magnetic stan...
Embodiment 3
[0103] Example 3 The one-step method of enzyme digestion and ligation is used for the assembly of different numbers of fragments
[0104] For the insert fragments encoding different lengths, use the one-step enzyme digestion ligation system to assemble the fragments on the approximate model plasmid, and check the success probability of the connection by colony PCR and sequencing. The specific experimental procedure is as follows.
[0105] 1. Design insert fragments of different lengths, and the sequence information is shown in Table 12.
[0106] Table 12 Fragment primer information for 2 or 4 fragment ligation
[0107]
[0108] 2. Configure the amplification system according to Table 8, perform amplification according to the procedures in Table 9, and purify the fragments according to the operation in Example 2.
[0109] 3. According to Table 13, configure the connection system of the model carrier and the fragment.
[0110] Table 13. Multi-fragment linkage system
[01...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com