Crohn disease biomarker, kit and screening method of biomarker
A biomarker, Crohn's disease technology, applied in the field of Crohn's disease biomarkers, kits and biomarker screening, can solve problems such as staying in, and achieve accurate results, high accuracy, and specificity high effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
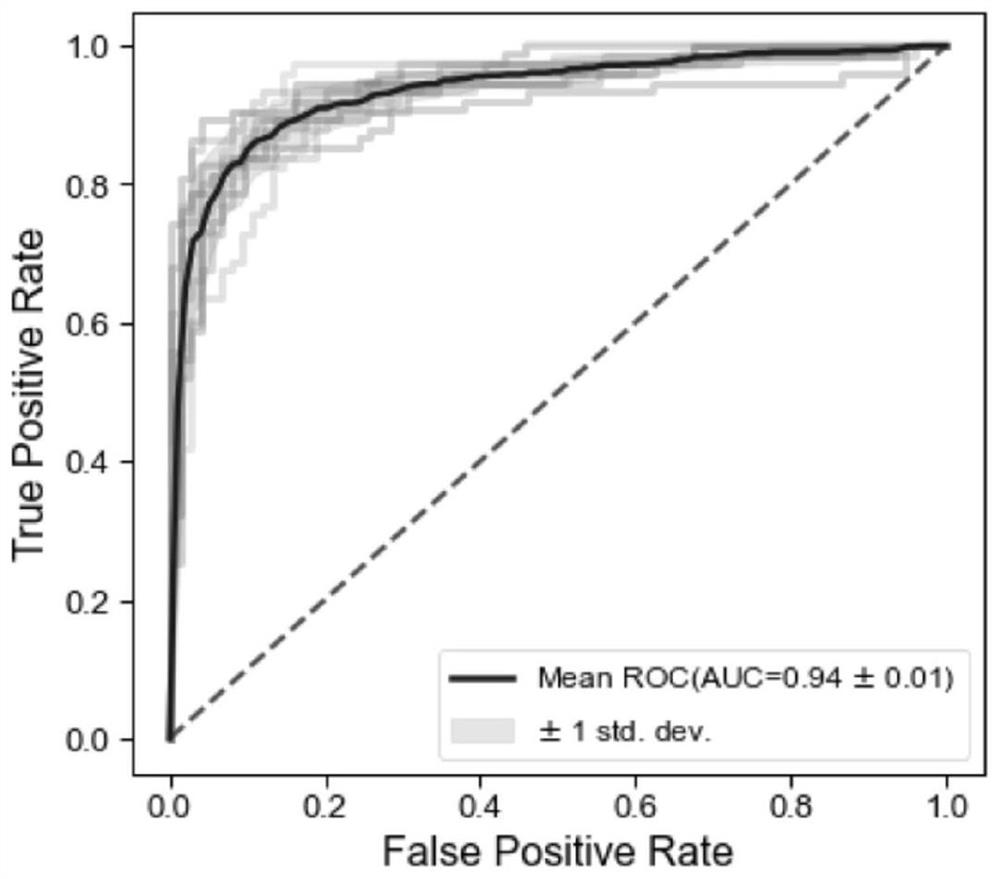
[0071] Example 1 Screening of markers for non-invasive diagnosis of Crohn's disease based on microbial homologous genes and construction of diagnostic models
[0072] 1.1. Data collection
[0073] From the National Institutes of Health Human Microbiome Project IBDMDB database (URL: https: / / ibdmdb.org), the US National Center for Biotechnology Information SRA database (URL: https: / / www.ncbi.nlm.nih.gov / sra ) and the European Bioinformatics Institute ENA database (website: https: / / www.ebi.ac.uk / ena) to obtain fecal microbial metagenomic sequencing data and clinical information data (clinical information Mainly including: disease status, age, sex and BMI).
[0074] The cohorts included in this example are: PRJNA398089, PRJNA389280, PRJNA400072, PRJNA385949, SRP057027; the number of samples included in the actual analysis is 1148, including 745 samples of Crohn's disease and 403 healthy controls.
[0075] 1.2. Data preprocessing
[0076] Quality control of the sequencing data w...
Embodiment 2
[0091] Example 2 Different cohort cross-validation and leave-one-out method validation
[0092] Experimental materials: Cross-validation and leave-one-out validation using public data from different cohorts to test the robustness and versatility of microbial biomarkers.
[0093] experimental method:
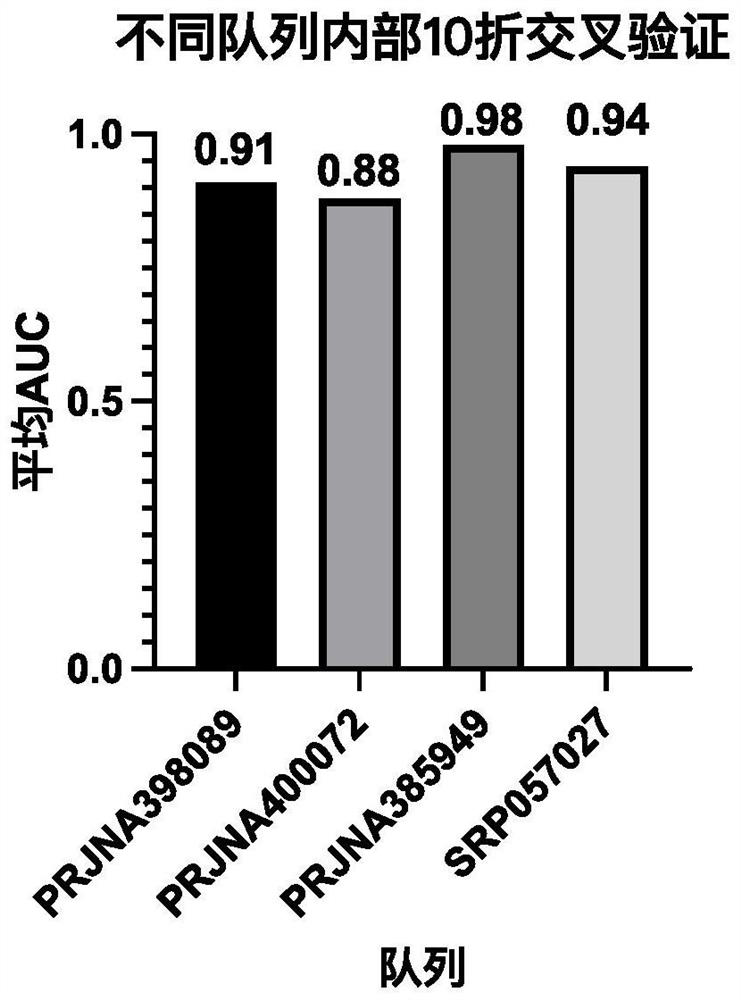
[0094] 2.1, 10-fold cross-validation within different cohorts
[0095]For public data from different cohorts, based on our identified optimal microbial homologous gene combinations (celB, nirK, lpxR, dadA, cshB, impH, manZ, ABCC-BAC, K03710, comFA, actP, E2.4.1.5, C5AP, thiK, ezrA, truC, czcA, a total of 17 homologous genes), carry out internal 10-fold cross-validation for each cohort, that is, each cohort is randomly divided into 10 folds, each fold is used as a test set in turn, and the remaining The 9 folds are used as the training set for model building, and the average AUC of 10 folds is obtained.
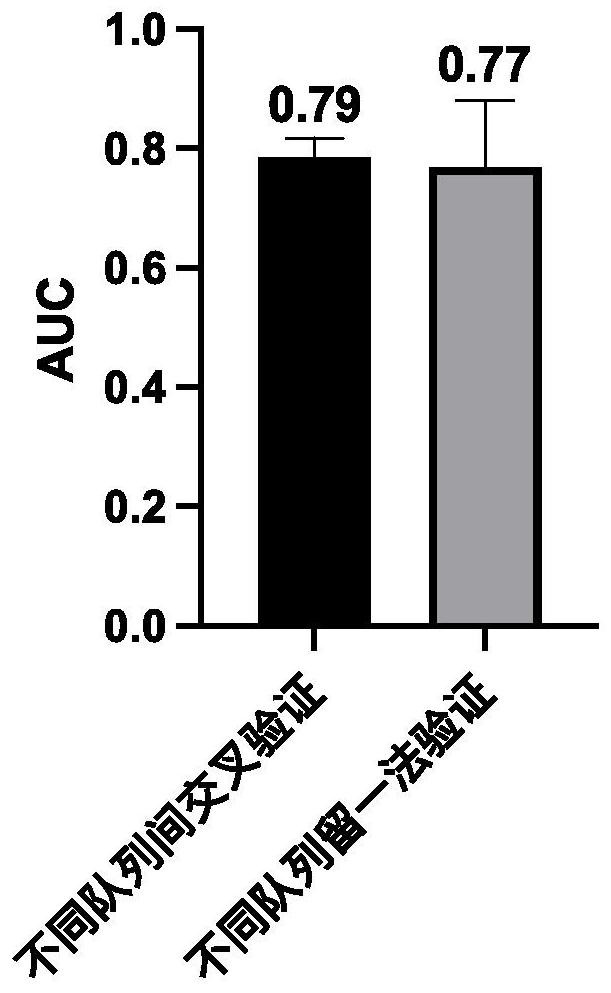
[0096] 2.2 Cross-validation between different cohorts
[0097] For the p...
Embodiment 3
[0100] Example 3 specificity verification
[0101] Experimental materials: Collect the sequencing data of intestinal disease microorganisms other than Crohn's disease in the database for specificity verification, including colorectal cancer (PRJEB27928, the number of disease samples is 22, and the number of healthy control samples is 60), Alzheimer's Momo (cohort PRJEB17784, the number of disease samples is 30, the number of healthy control samples is 28), type 2 diabetes (cohort PRJEB1786, the number of disease samples is 53, the number of healthy control samples is 43) and liver cirrhosis (cohort PRJEB6337, the number of disease samples is 126, and the number of healthy control samples was 94).
[0102] Experimental method: For the sequencing data of different diseases, based on the optimal combination of homologous gene markers confirmed by us, a model was constructed for each disease, and the result of 10-fold cross-validation was obtained, that is, each disease data was r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com