SgRNA for improving drought tolerance and saline-alkaline tolerance of cotton and application of sgRNA
A technology for salt and alkali tolerance and cotton, applied in the field of sgRNA, can solve the problems of undiscovered cotton drought tolerance and salt and alkali tolerance, and achieve the effects of high gene editing efficiency, improved salt and alkali tolerance, and improved cotton fiber quality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The design of embodiment 1 cotton Gigantea (GI) gene target sequence
[0045] (1) GI gene sequence acquisition
[0046] Using the cotton FGD database ( https: / / cottonfgd.org / ) searched for the cotton Gigantea (GI) gene, and found four GI homologous genes, which were located on chromosomes D02, D05, A02, and A05, respectively. The 4 GI homologous genes were compared and searched in the NCBI online database, and a total of 16 accession numbers were found, including D02 chromosome genes XM_016838310.2, XM_041088410.1; D05 chromosome genes XM_041094387.1, XM_041094386.1, XM_041094385.1, XM_041094384.1,XM_041094383.1,XM_041094382.1;A02染色体基因XM_041090781.1,XM_016886709.2,XM_016886708.2;A05染色体基因XM_016842825.2,XM_041112076.1,XM_016842823.2,XM_016842822.2,XM_041112077.1。
[0047] (2) GI gene target site design rules
[0048] Using the GI gene ID number obtained in step (1), download the nucleotide sequences of all GI genes, and perform comparative analysis to obtain their co...
Embodiment 2
[0053] Example 2 Construction of CRISPR / Cas9 Gene Editing Expression Vector
[0054] Proceed as follows:
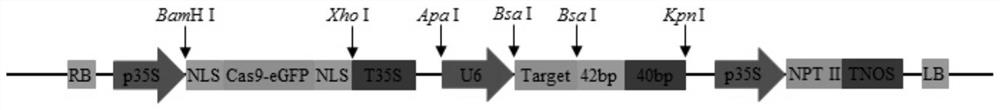
[0055] (1), construction of pGBI-Cas9 expression vector: Cas9 gene and eGFP gene are fused, and a nuclear localization signal (NLS) sequence is added at the 5' and 3' ends of the fusion gene to obtain the Cas9-eGFP fusion gene; then the fusion The gene fragment was connected to the pBI121 expression vector cut by BamH I and Xho I to obtain a plant expression vector named pGBI-Cas9. The expression vector was regulated by a 35S promoter and a 35S terminator to express the Cas9-eGFP fusion gene. The reaction system is: fusion gene 1μL (10ng), pBI121 vector 2μL (20ng), T4 DNA ligase (NEB) 1μL, T4 DNA ligase Buffer 1μL, ddH 2 05 μL. Ligated overnight at 16°C to obtain the ligated product; then the ligated product was transformed into Escherichia coli and screened by sequencing to obtain the plant expression vector pGBI-Cas9 with the correct Cas9-eGFP fusion gene sequence.
...
Embodiment 3
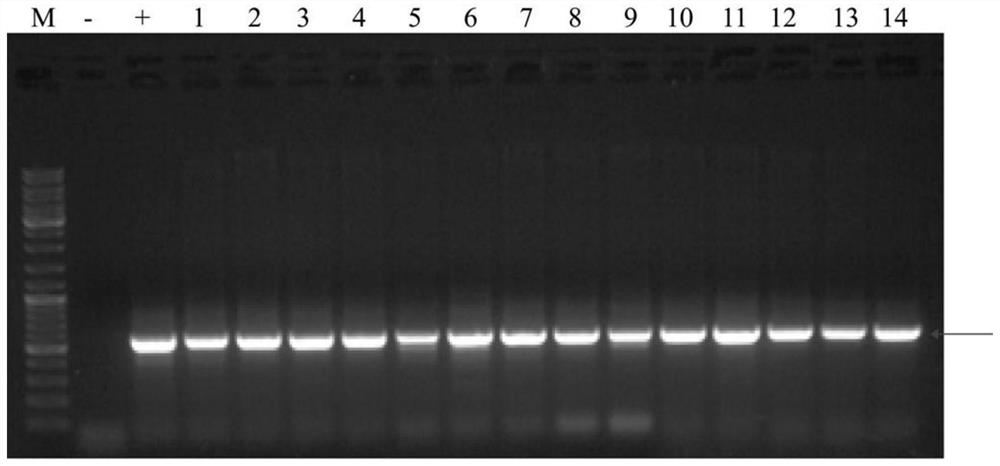
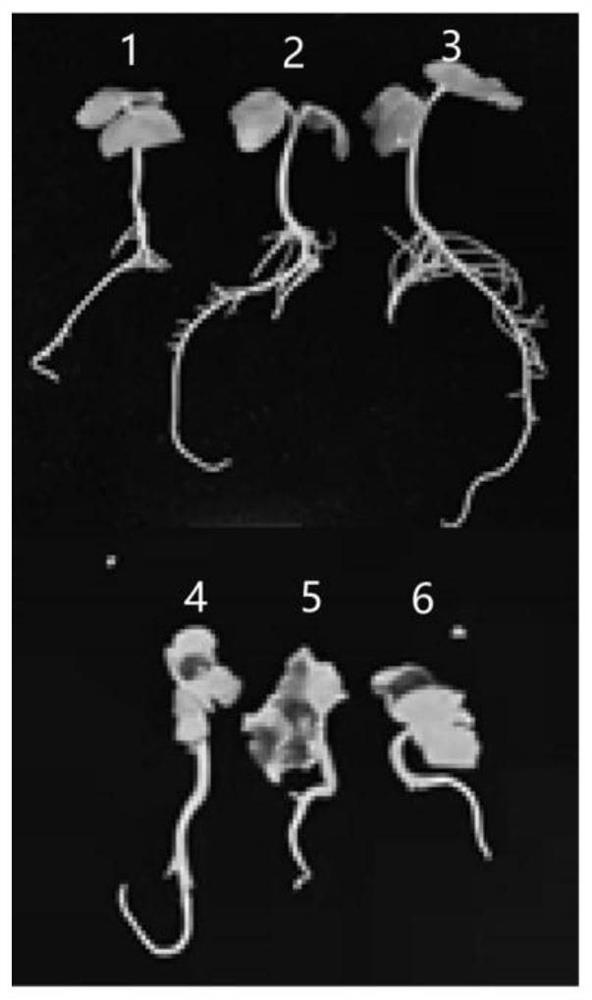
[0063] Example 3 The acquisition of cotton GI gene knockout mutant material
[0064] (1) Gene editing expression vector pGBI-Cas9-GI-KO transformed into Agrobacterium
[0065]Mix 100 μL of Agrobacterium GV3101 competent cells with 2-5 μg of the gene editing expression vector pGBI-Cas9-GI-KO obtained in Example 2 to obtain a suspension mixture, and place it on ice for 5 minutes. Add the suspension mixture to a 2mm electric shock cup (try to avoid generating air bubbles when adding it), and electric shock at 2500V; quickly add 800 μL of LB liquid medium after electric shock and transfer it to a 1.5mL centrifuge tube, activate and culture at 28°C and 180rpm for 5 hours, and obtain Agrobacterium suspension containing pGBI-Cas9-GI-KO expression vector. 200 μL of bacterial liquid was taken out and spread on LB solid medium plate containing spectinomycin and rifampicin (both at a concentration of 50 μg / mL), and cultured at 28° C. for 48 h. Since the pGBI-Cas9-GI-KO expression vecto...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com