Standard plasmid for detecting copy number of exogenous gene of recombinant CVA10 vaccine as well as preparation method and application of standard plasmid
A CVA10 and foreign gene technology is applied in the field of standard plasmids for detecting the copy number of exogenous genes of recombinant CVA10 vaccine, and achieves the effects of saving detection time, simple operation and shortening detection time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0083] 1.2. Preparation of recombinant plasmids
[0084] 1.2.1. The tandem CVA10-MOX-P1-3CD product was obtained by PCR
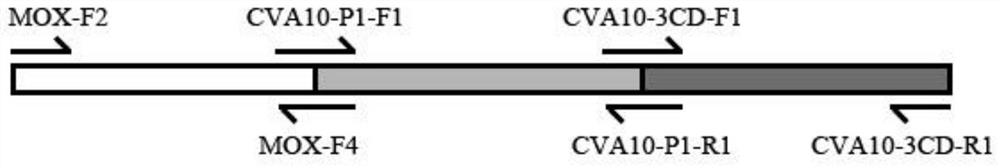
[0085] 1. Using the Hansenula yeast genome as the template and MOX-F2 and MOX-R3 as primers, PCR amplification was performed to obtain the endogenous gene MOX (SEQ ID No.1);
[0086] ②. Using the recombinant CVA10 vaccine Hansenula genome as the template, and using CVA10-P1-F1 and CVA10-P1-R1, CVA10-3CD-F1 and CVA10-3CD-R1 as primers, PCR amplification was performed to obtain foreign genes P1 fragment (SEQ ID No. 2) and exogenous gene 3CD fragment (SEQ ID No. 3);
[0087] ③. Establish the PCR reaction system shown in Table 1 below. The PCR amplification in Steps ① and ② is performed according to the PCR reaction system in Table 1.
[0088] Table 1 PCR reaction system
[0089]
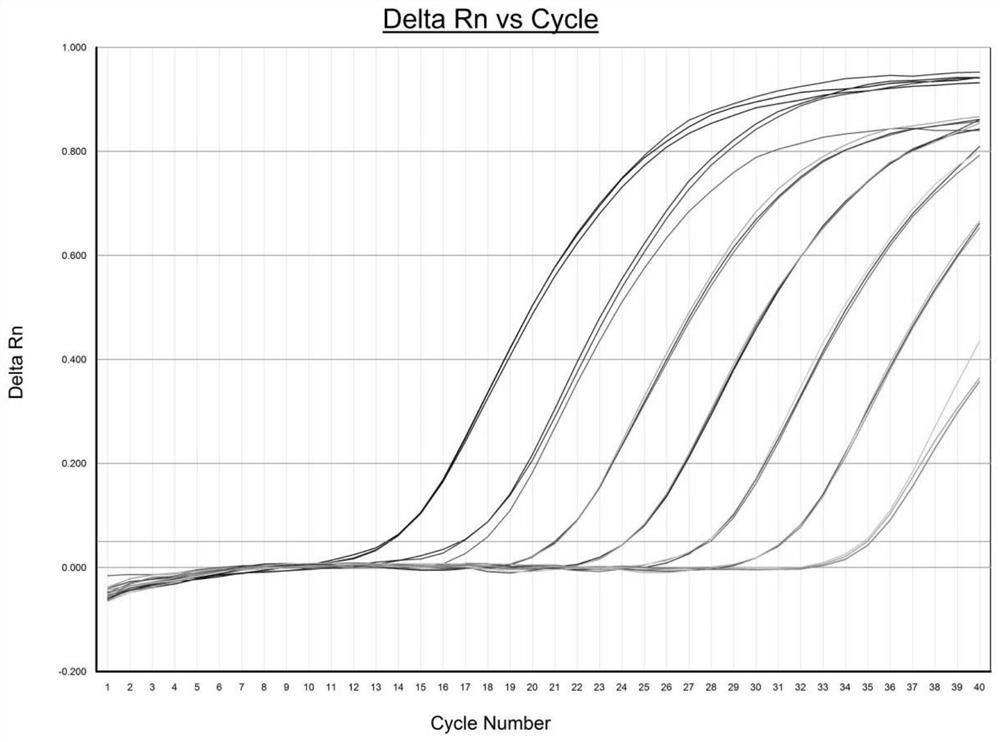
[0090] ④. The PCR amplification of endogenous gene MOX, exogenous gene P1 and exogenous gene 3CD was detected by 1% agarose electrophoresis. The sizes of the corresponding PCR ...
Embodiment 2
[0160] Example 2: The difference from Example 1 is that the cloning vector in this example is the pUC18 vector, and the competent cells are TOP10 competent cells.
Embodiment 3
[0161] Example 3: The difference from Example 1 is that the cloning vector in this example is the pUC18 vector, and the competent cells are JM109 competent cells.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com