Polypeptide PROTAC molecule as well as preparation method and application thereof
A molecular and reaction unit technology, applied in the field of polypeptide PROTAC molecules and their preparation, can solve the problems of not being able to bring the target protein and E3 ligase closer, and limiting the clinical transformation of PROTAC molecules.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
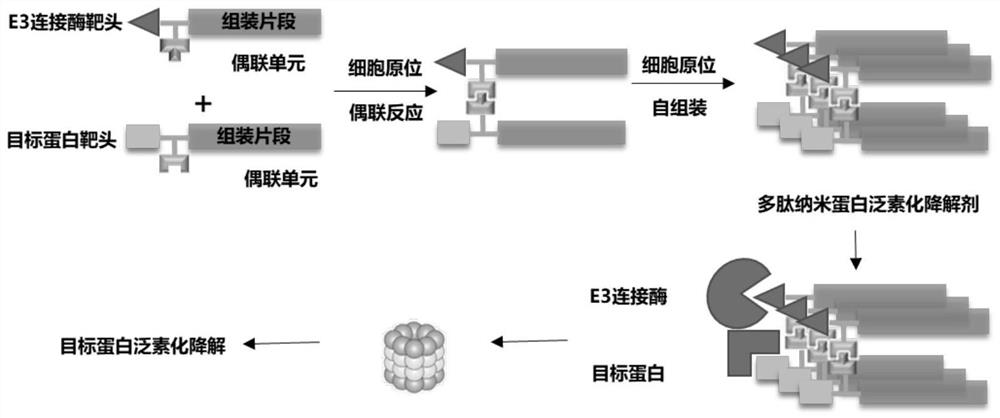
[0151] This embodiment provides a polypeptide nanoprotein ubiquitination degradation agent, which can activate and catalyze the click reaction process under the reduction of GSH in tumor cells, and trigger the assembly process at the same time. A nanofibrous polypeptide nanoprotein ubiquitination degradation agent is formed, and the formation process schematic diagram of the nanofibrous polypeptide nanoprotein ubiquitination degradation agent is as follows figure 1 shown.
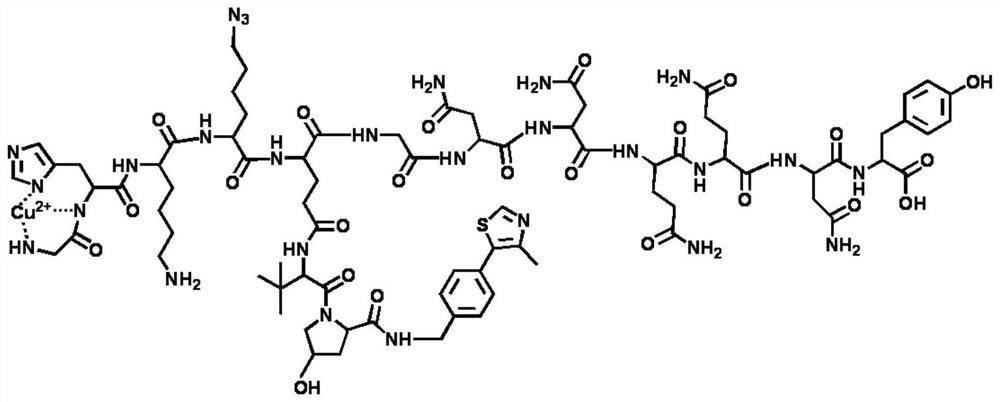
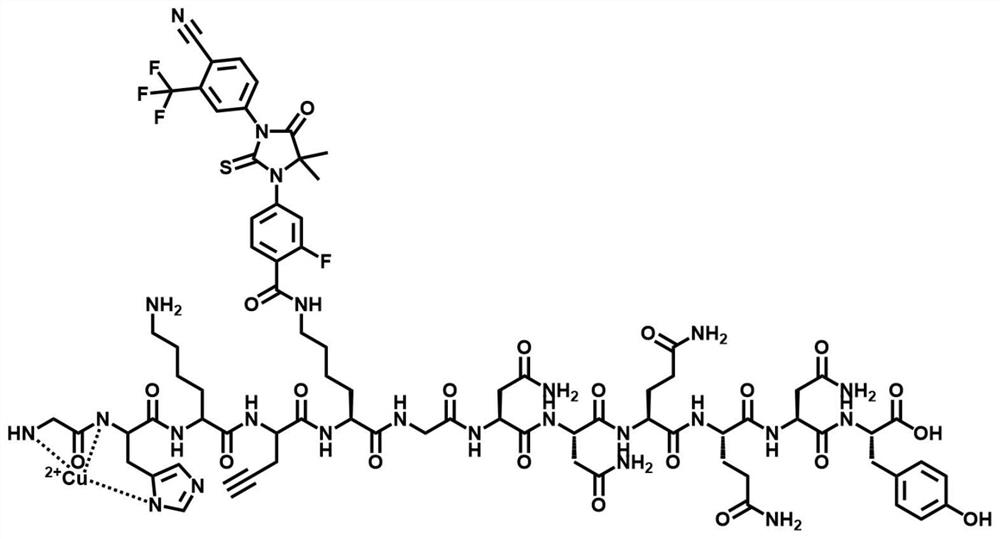
[0152] The module molecule A of the polypeptide nanoprotein ubiquitination degradation agent targets and recognizes E3 ligase VHL Ligand 1, and the module molecule B targets and recognizes AR; the module molecule A is GHK (Cu 2+ )K(N 3 )-E-(VHL-1)GNNQQNY, the structure of the module molecule A is as figure 2 Shown; The module molecule B is GHK (Cu 2+ ) Pra-K-(Enza)GNNQQNY, the structure of the module molecule B is as image 3 shown.
[0153] In module molecule A and module molecule B, in GHK, G (glyci...
Embodiment 2
[0168] This embodiment provides a polypeptide nanoprotein ubiquitination degradation agent, the module molecule A of the polypeptide nanoprotein ubiquitination degradation agent is consistent with the module molecule A in Example 1, and the module molecule B targets and recognizes EGFR; Module molecule A is GHK(Cu 2+ )K(N 3 )-E-(VHL-1)GNNQQNY; the module molecule B is GHK(Cu 2+ ) Pra-K-(Gefi)GNNQQNY, the structure of the module molecule B is as Figure 4 Shown; In the module molecule B, Gefi represents a gefitinib derivative. The preparation method of the polypeptide nanoprotein ubiquitination degradation agent refers to the preparation method of Example 1.
Embodiment 3
[0170] This embodiment provides a polypeptide nanoprotein ubiquitination degradation agent, the module molecule A of the polypeptide nanoprotein ubiquitination degradation agent targets and recognizes the E3 ligase CRBN, and the module molecule B is consistent with the module molecule B in Example 1 ; The module molecule A is GHK (Cu 2+ )K(N 3 )-E-(CRBN)GNNQQNY, the structure of the module molecule A is as Figure 5 shown.
[0171] In module molecule A, CRBN represents a targeting molecule targeting E3 ligase CRBN. The preparation method of the polypeptide nanoprotein ubiquitination degradation agent refers to the preparation method of Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com