Helicase activity determination method based on double-stranded DNA and application of helicase activity determination method
A helicase and activity technology, applied in the field of enzyme activity detection, can solve the problems of limited expansion DNA substrate model design, cumbersome detection methods, fluorescence quenching, etc., and achieve accurate quantitative effect of helicase activity and helicase activity. The effect of comprehensive evaluation and shortened detection time of
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0132] In order to make the invention purpose, technical solutions and technical effects of the present invention clearer, the present invention will be further described in detail below with reference to the specific embodiments. It should be understood that the specific embodiments described in this specification are only for explaining the present invention, rather than for limiting the present invention.
[0133] The experimental materials and reagents used, unless otherwise specified, are conventional consumables and reagents that can be obtained from commercial sources.
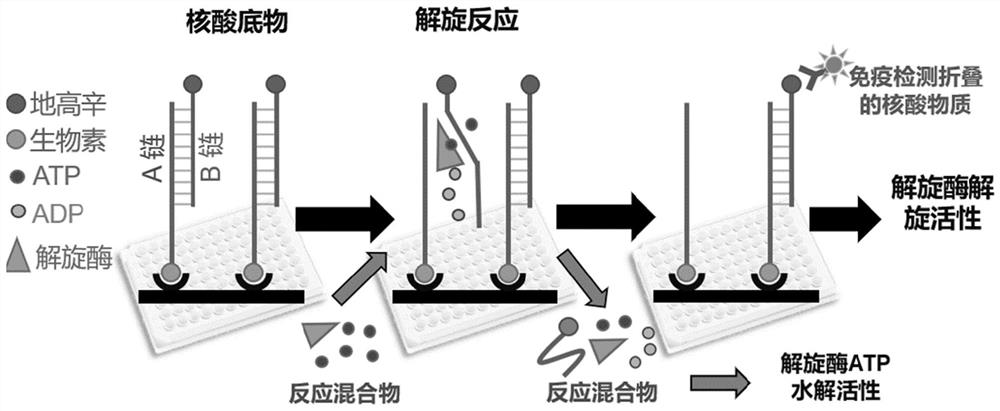
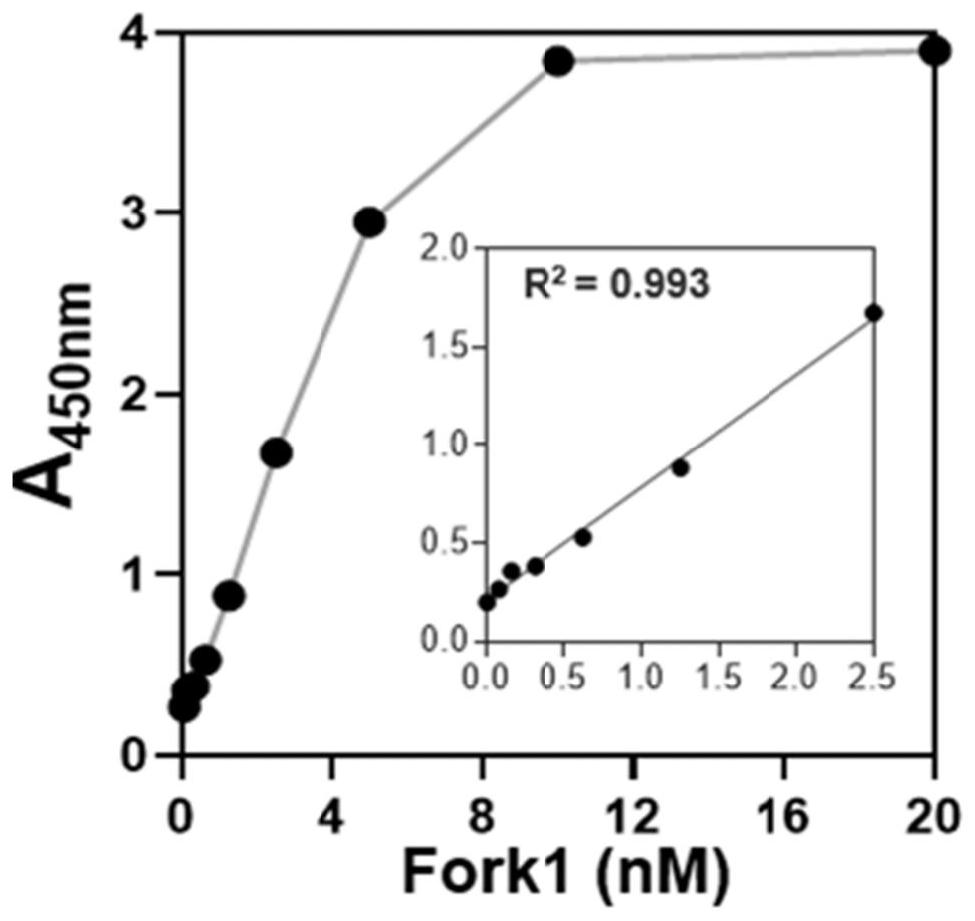
[0134] A method for measuring helicase activity based on double-stranded DNA
[0135] The double-stranded DNA-based helicase activity assay method in this embodiment includes the following steps:
[0136] (1) Design of double-stranded DNA:
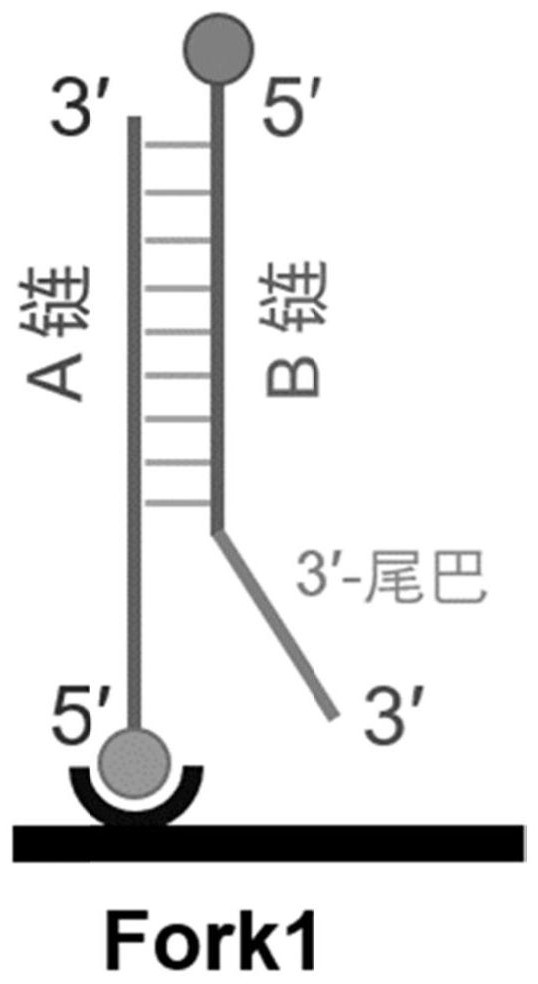
[0137] In this example, the double-stranded DNA used is a replication fork double-stranded DNA.
[0138] The replication fork (fork1) was selected from a bifurca...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com