Method for knocking out mouse Abhd12 gene by using CRISPR/Cas9 system and application
A systematic knockout and mouse technology, applied in the field of transgenics, to reduce the possibility of off-target and improve specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
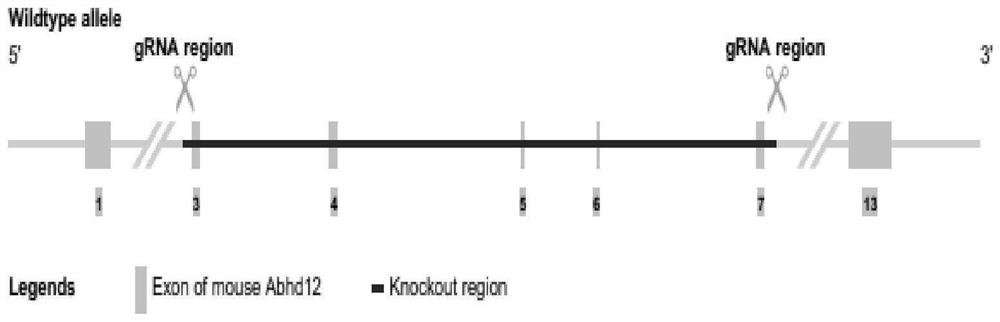
[0035] Synthesis of targeted knockout gene sgRNA: see appendix for knockout strategy figure 1 .
[0036] The nucleotide sequence of the mouse Abhd12 gene was downloaded from the NCBI database and homologous alignment was performed. According to the PAM design principle of guideRNA and the results of homology alignment, crRNA1 and crRNA2 were synthesized, and the sequences are:
[0037] 1. Synthetic crRNA1 sequence
[0038] SEQ ID NO.1=5'-GGCACAGAGGTTCACGTTTCGUUUUAGAGCUAUGCUGUUUUG-3';
[0039] 2. Synthetic crRNA2 sequence
[0040] SEQ ID No. 2 = 5'-GTAGCACTTGTCATCTACCAGUUUUAGAGCUAUGCUGUUUUG-3'.
Embodiment 2
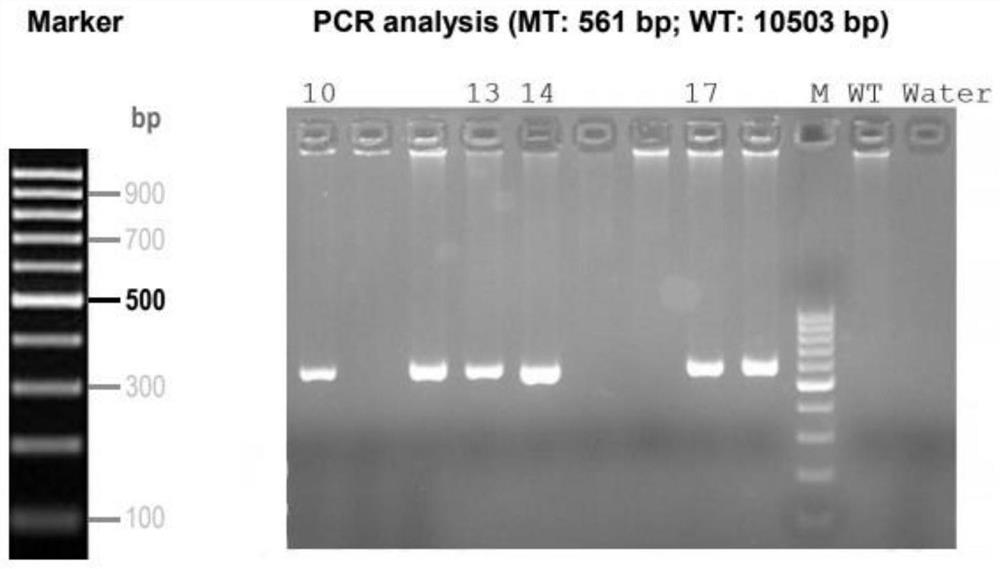
[0042] C57BL / 6 female mice were treated with PMSG, then injected with hCG, and mated with male mice. The fertilized eggs were collected the next day for microinjection. Item No. M0646)]: co-injected into fertilized eggs, take the fertilized eggs that survived the injection and transplant them into pseudopregnant female mice, the embryo-transferred mice will be born about 21 days after the operation, that is, the F0 generation mice. 14 days after birth, the mice were tail clipped to extract DNA and identified by PCR. The time from DNA extraction to PCR detection is 2-3 days. So this cycle takes about 40 days. The results of PCR identification of F0 mice are shown in the attachment. figure 2 .
Embodiment 3
[0044] When male Founder mice are 8 weeks old and female mice are 6 weeks old, they can be mated with wild-type heterozygous mice to obtain F1 generation heterozygous mice. The mice are identified by PCR 14 days after birth. If positive mice are born, It means that the transgene has been integrated into the germ cells, and this process takes about 120 days.
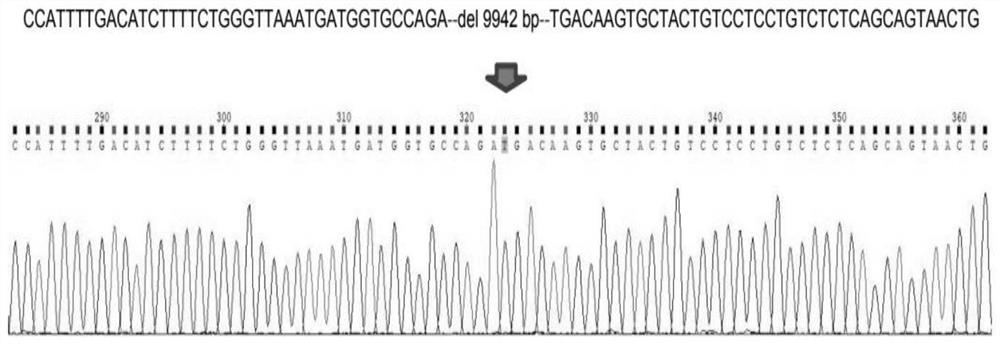
[0045] attached image 3 For the positive mouse sequencing analysis, it can be seen that 9942bp has been deleted between the genomes;
[0046] attached Figure 4 For the identification of F1 mice, mice 26, 27, 28, 35, 36 and 37 were identified as positive F1.
[0047] PCR primer F1 is designed on the upstream outside of the knockout region, R1 is designed on the downstream outside of the knockout region, and R2 is designed in the knockout region; appendix Figure 5 Relative position of PCR primers on the genome.
[0048] The primer sequences are as follows
[0049] PCR Primers1 (Annealing Temperature 60.0℃):
[0050...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com