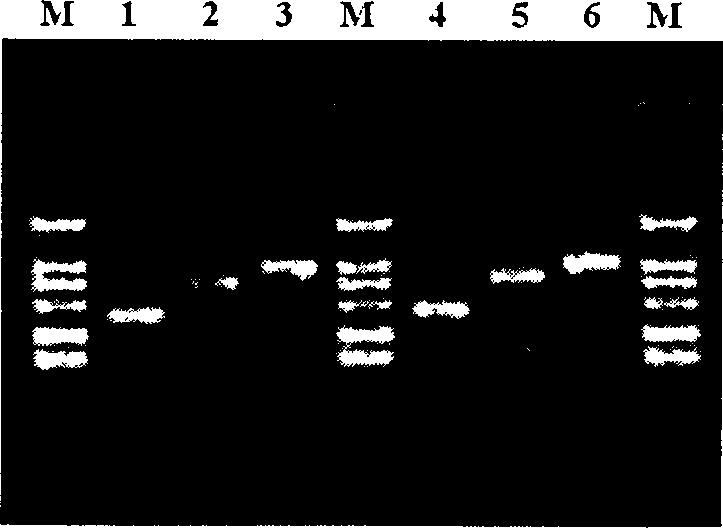Method for preparing nucleic acid probe library
A technology of nucleic acid probes and probes, which is applied in biochemical equipment and methods, microbial measurement/inspection, fermentation, etc., and can solve the problem of inability to analyze or discover unknown genes, probe libraries that have not been widely used, and disposable High purchase cost and other issues, to achieve the effect of strong anti-strike ability, reduce dependence, and convenient transportation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
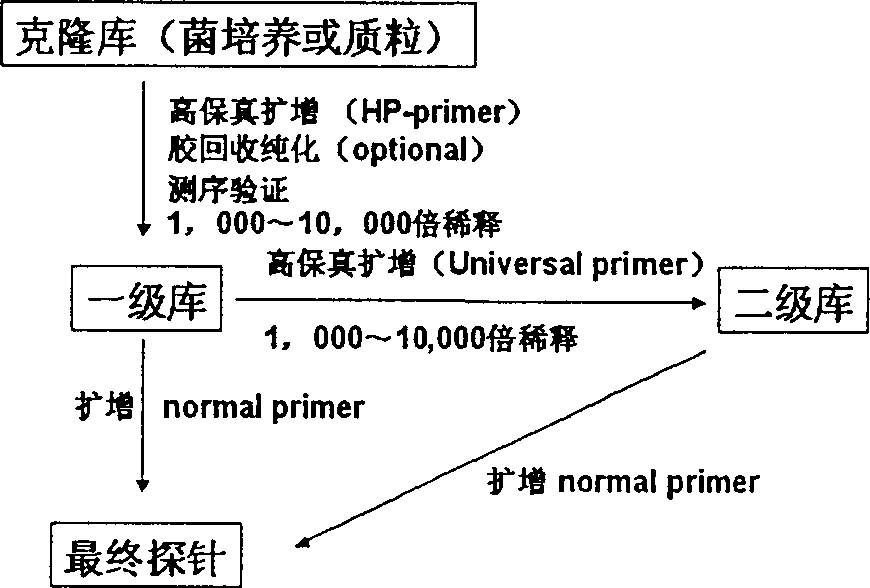
Image
Examples
Embodiment 1
[0017] Example 1: Preparation of Human Gene Cloning Probe Library
[0018] 1. Human gene probe cloning
[0019] Three clones 175C1, 175D3, and 175A5 of different lengths were randomly selected from the human gene clone library of the Beijing National Engineering Research Center for Biochips (the uniform number of the National Engineering Research Center for biochips), and the lengths of the human gene clones contained in them were respectively 490bp, 750bp and 1000bp, basically covering all clones in length. These clones will serve as templates for profiling microarray probe preparation in two different formats. The first form is preserved engineering bacteria culture, and the second form is plasmid prepared by alkaline lysis method.
[0020] 2. Primers
[0021] Conventional primer: upstream primer 5'CTG CAA GGC GAT TAA GTT GGG TAA C 3';
[0022] Downstream primer 5'GTG AGC GGA TAA CAA TTT CAC ACA GGA AAC AGC 3';
[0023] Universal primer: upstream primer 5'TCA ...
Embodiment 2
[0038] Example 2. Preparation of clinical infectious disease pathogen clone probe library
[0039] 1. Cloning of pathogenic probes for clinical infectious diseases
[0040] Four clones pTP205, pHCV244, pHIV309 and pHBV538 with different lengths were selected from the clinical infectious disease pathogen clone library of Beijing National Engineering Research Center of Biochip, and the lengths of the clinical infectious disease pathogen clones contained in them were 205bp and 244bp respectively , 309bp and 538bp. These clones correspond to four different pathogens: TP (Treponema pallidum), HCV (Hepatitis C virus), HIV (AIDS virus) and HBV (Hepatitis B virus). These clones will be in the form of plasmids as templates for microarray probe preparation.
[0041] 2. Primers
[0042] Conventional primers: Treponema pallidum: upstream primer 5'AAC ACG ATC CGC TAC GAC TAC TAC 3',
[0043] Downstream primer 5'CCC TAT ACC CGT TCG CAA TCA AAG 3';
[0044] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com