Nucleic acid molecular hybridization detection method for silkworm nosema disease
A technology of nucleic acid molecular hybridization and silkworm microparticles, which is applied in the measurement/testing of microorganisms, biochemical equipment and methods, etc., can solve the problems of poor accuracy, low sensitivity, and long time consumption, and achieve the effect of good quality and high content
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0016] The specific operation steps of the present invention are as follows:
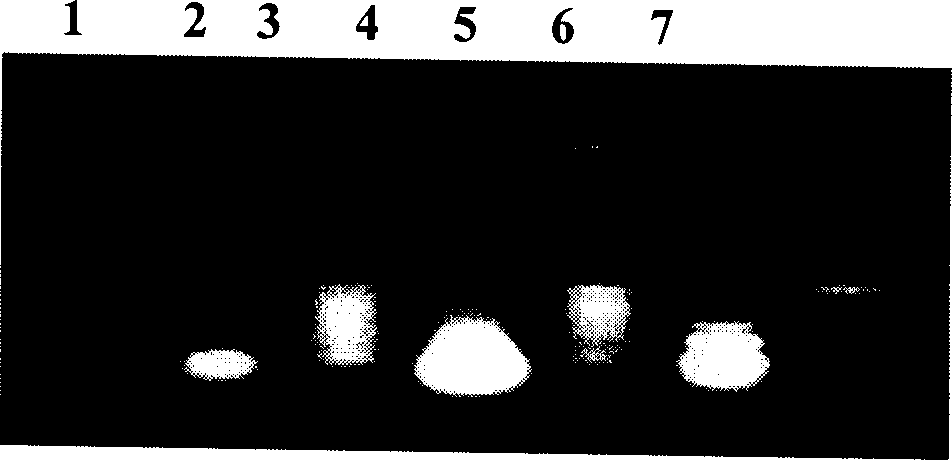
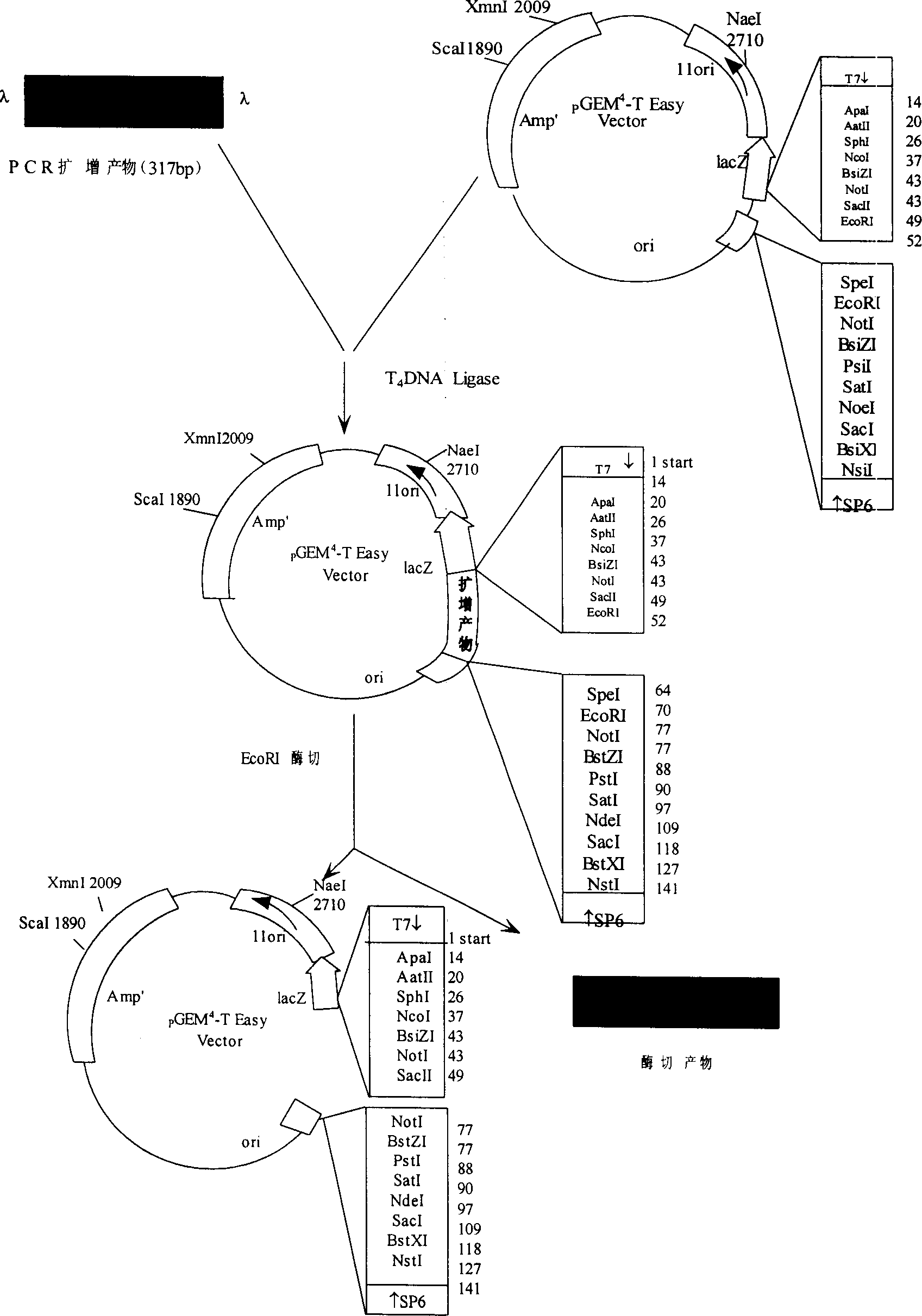
[0017] DNA extraction of Nosema bombycis → polymerase linked reaction (PCR) → PCR product cloning and recombinant plasmid identification → recombinant plasmid sequence analysis → DIG-labeled probe for specific identification of NB spores → DIG-labeled probe for detection of NB spore DNA Identification→Application of DIG-labeled probe in sericulture production.
[0018] 1. Extraction of Nosema bombycis DNA: Use proteinase K phenol chloroform extraction method to extract. To crush and purify the Bombyx mori, which is fed with microsporidia, is carried out as follows:
[0019] Take 400 microliters of centrifugation and remove the supernatant + equal volume of 0.2 mol / L K 2 CO 3
[0020] ↓30℃, 1 hour, remove impurities
[0021] Transfer to PBS buffer (pH 7.8)
[0022] ↓30℃, 2 hours, stimulate spore germination
[0023] 6000 rpm, 5 minutes, remove the supernatant, add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com