Protein expression profiling
A purpose and analyte technology, applied in the direction of analytical materials, microbial measurement/inspection, instruments, etc., can solve problems such as difficult to simplify operations, small size, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
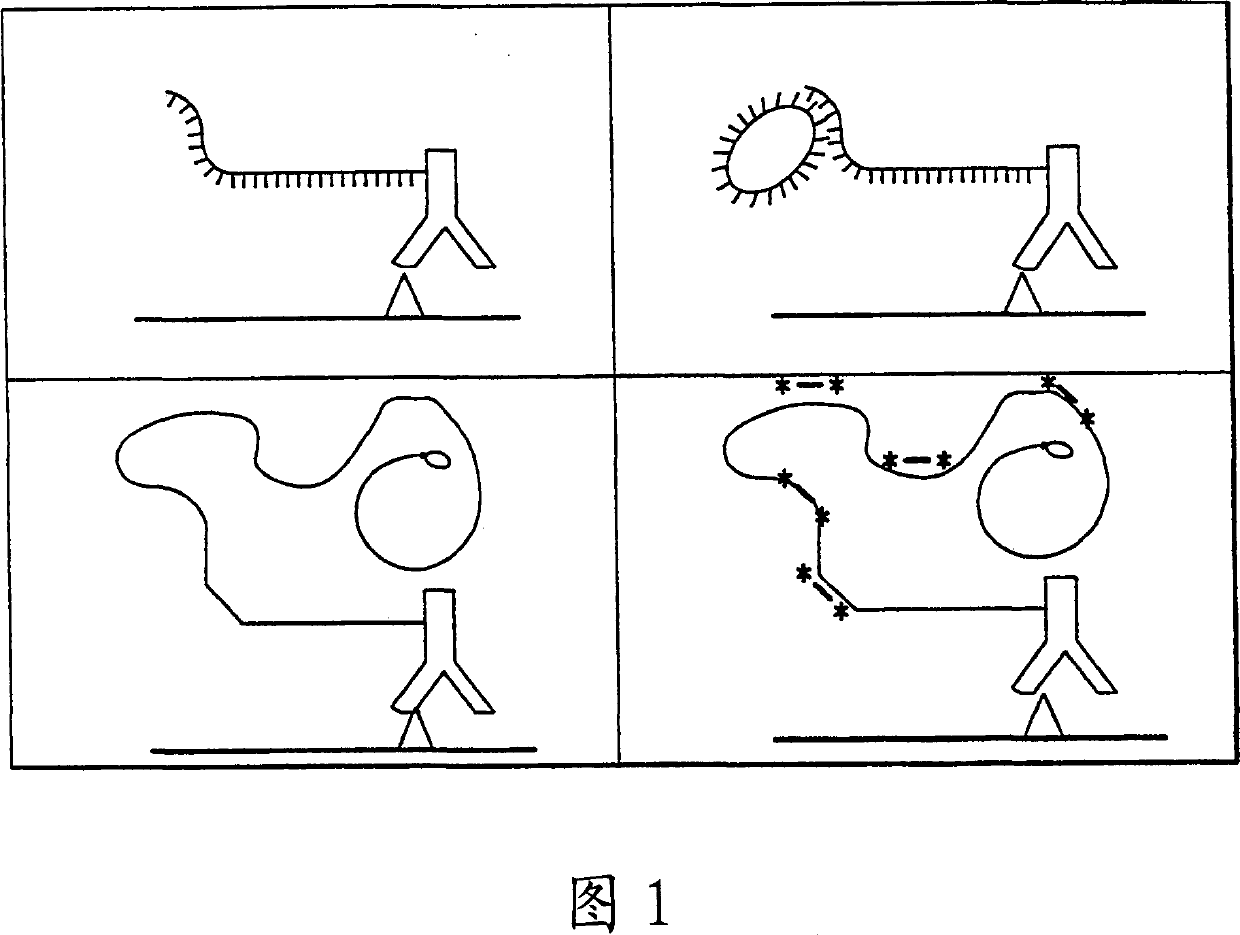
[0181] This example illustrates the construction and characterization of antibody DNA conjugates used as Reporter Binding Primers
[0182] Oligonucleotides. All oligonucleotides used were synthesized by Perseptive Biosystems Rapid DNS Synthesizer and purified by reverse phase HPLC. Circular DNA was constructed as previously described (4). Conjugate rolling circle replication primer: 5'-thiol-GTA CCA TCA TAT ATG TCC GTG CTA GAA GGA AACAGT TAC A-3'; amplified target circular DNA: 5'-TAG CAC GGA CAT ATA TGA TGG TAC CGCAGT ATG AGT ATC TCC TAT CAC TAC TAA GTG GAA GAA ATG TAA CTG TTTCCT TC-3'; Detection Probe-5'Cy3 TAT ATG ATG GTA CCG CAG Cy3 3', 5'Cy3TGA GTA TCT CCT ATG ACT Cy33', 5'Cy3 TAA GTG GAA GAA ATG TAA Cy33.
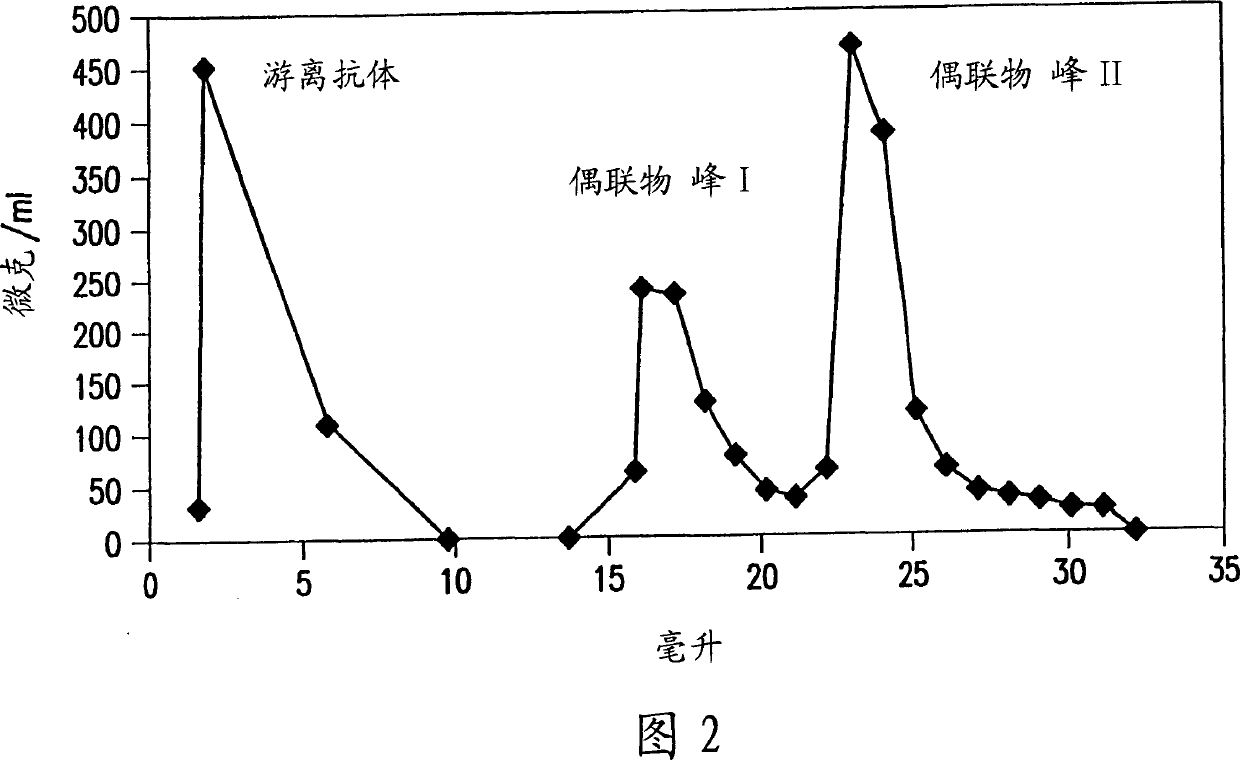
[0183] Antibody-DNA Conjugates. Antibodies were buffer exchanged into a buffer of 50 mM sodium phosphate pH 7.5, 150 mM NaCl, 1 mM EDTA by chromatography on a PD-10 column (Amersham-Pharmacia Biotech). Desalted antibody (41 nmoles) was treated with a 10-fold mola...
Embodiment 2
[0187] This example illustrates the use of indicator antibody primers / antibody-DNA conjugates for the detection of analytes in an ELISA assay. Compared with the use of conventional antibody-enzyme conjugates, the immuno-RCA assay showed high sensitivity and good kinetic range.
[0188] ELISA test. The 96-well plate was covered with goat polyclonal anti-human IgE at 100 μl 2 mg / ml per well for 2 hours at 37°C, then overnight at 4°C. The plate was washed 3 times with 100 μl TBS / 0.05% Tween20, and then blocked with 5% non-fat dry milk for 2 hours at 37°C. Plates were washed again with TBS / 0.05% Tween20 before addition of IgE analyte at various concentrations in a volume of 100 μl. After incubation at 37° C. for 30 min, the plate was washed 3 times with 100 μl TBS / 0.05% Tween20. In a conventional ELISA assay, anti-human IgE-alkaline phosphatase conjugate was added to each well and incubated at 37°C for 30min. After washing the plate with TBS / 0.05% Tween20, the alkaline phospha...
Embodiment 3
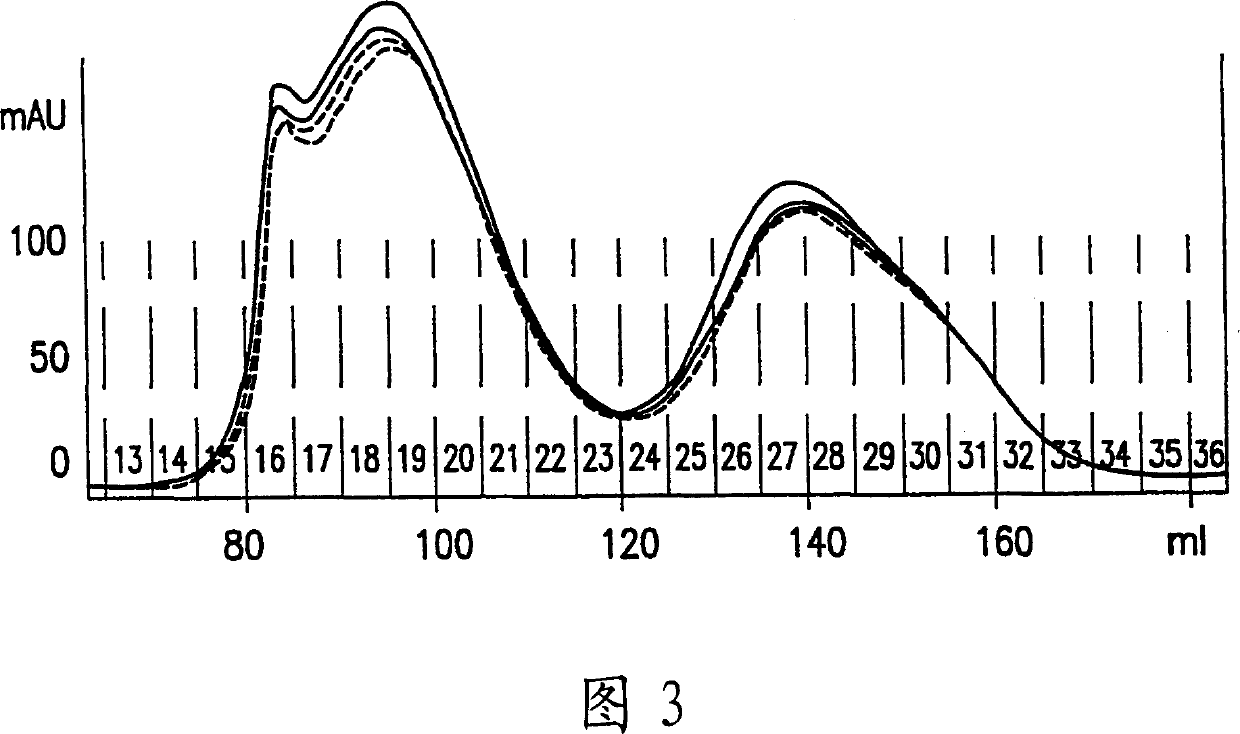
[0190] This example illustrates the use of indicator antibody primers / antibody-DNA conjugates to detect analytes in microparticle form. Human IgE was still chosen as the test analyte, but this sandwich assay was performed using avidin-coated magnetized microparticles and a biotinylated polyclonal anti-human IgE capture antibody. In general, streptavidin-coated magnetized beads (Bangs Laboratories) were covered with a solution of 16 μg / ml biotinylated polyclonal anti-human IgE (Pharmingen) in TBS and washed with TBS / 0.05% Tween20 for 3 Once, block overnight with 2mg / ml BSA. The beads were incubated with human IgE (25 ng / ml) for 20 min at room temperature in TBS and washed 3 times with TBS / 0.05% Tween20. IgE detection using conventional anti-IgE-DNA conjugates or immuno-RCA conjugates was performed as described above for the ELISA assay. In Figure 7, it can be seen that in the presence of moderate concentrations of IgE coupled to microparticles, immuno-RCA with anti-IgE-DNA co...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com