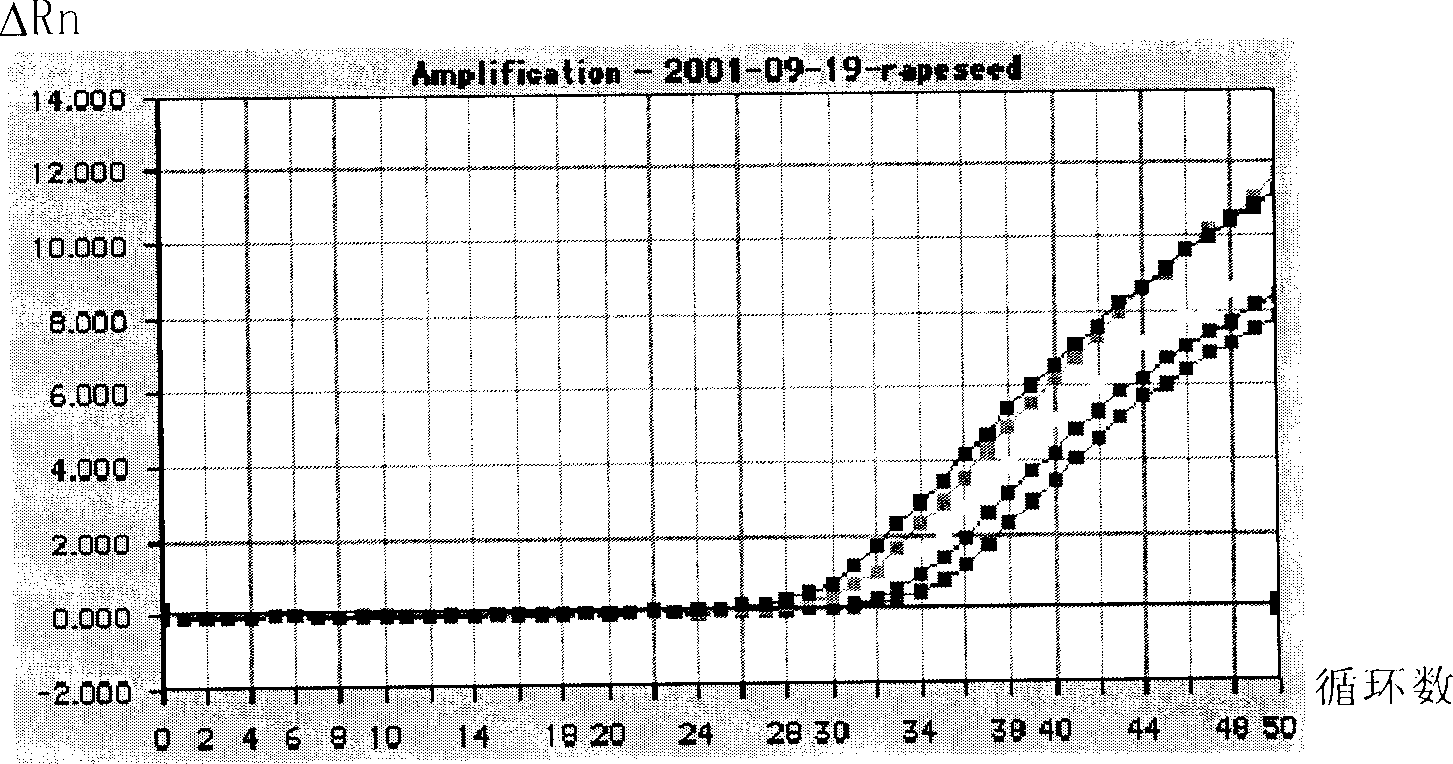
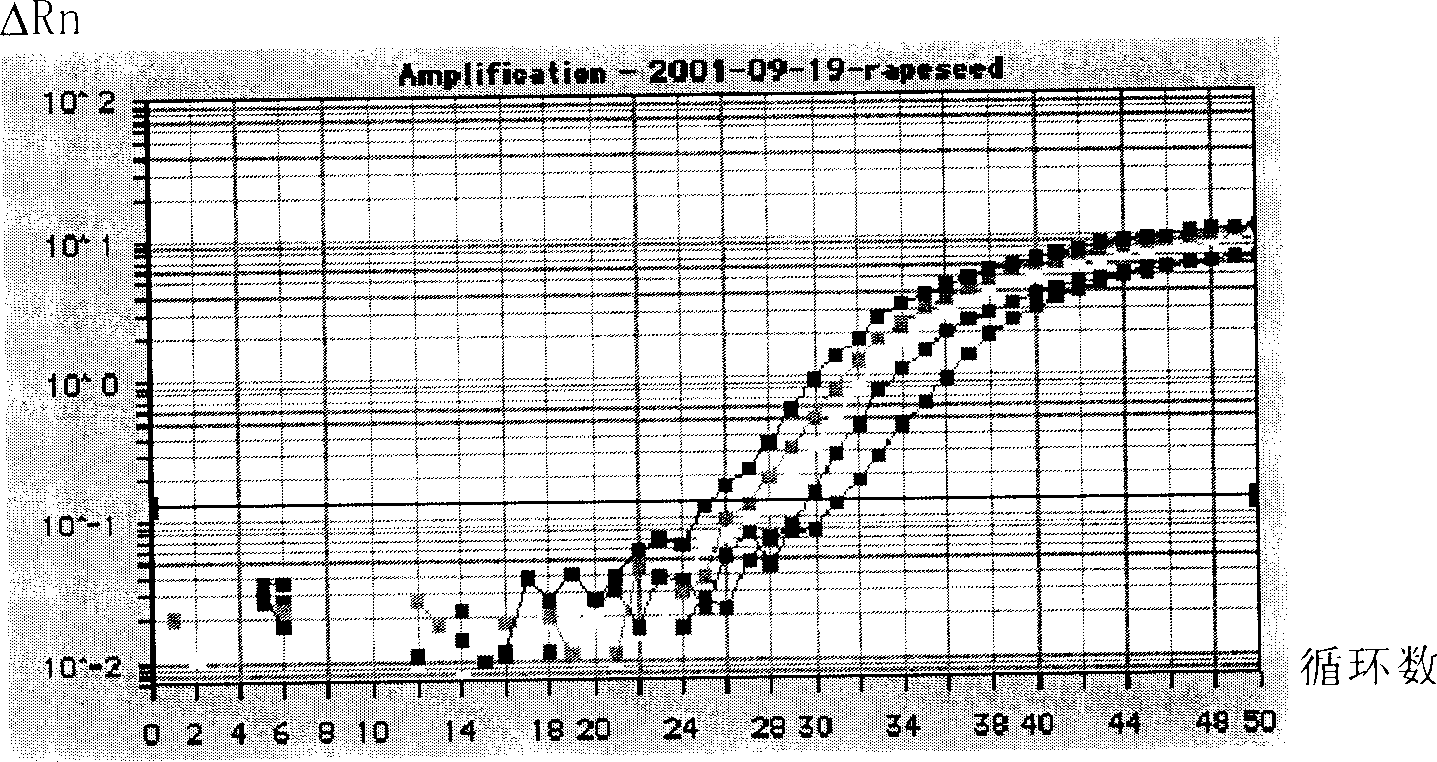
Quantitative measuring transgene component in transgene rapeseed and processed product
A technology for processing products, rapeseed, applied in the field of product testing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0144] Determination of Rapeseed Specific Internal Standard Gene
[0145] As an endogenous specific marker gene, the following two conditions need to be met: (1) the gene exists in all varieties of the crop; (2) other organisms do not have the gene, or there is no specific target sequence for amplifying the gene. The inventor found that the phosphoenolpyruvate carboxylase (Phosphoenolpyruvate-Carboxylase, PE3-PEPCase, PEP) gene can meet the above two conditions by repeatedly searching in GenBank, and the phosphoenolpyruvate carboxylase expressed by the gene is Catase is a key enzyme in the Calvin cycle of photosynthesis. The number of this gene in GenBank is: D13987. Through the Blast function of GenBank to find the homologous gene of this gene, it is found that this gene has a high specificity. Other genes There is very little homology to this gene.
Embodiment 2
[0147] Specificity analysis of PCR primers and products
[0148] According to the comparative analysis of the homology of the PEP gene, the inventors selected the sequence design primers of the 3626bp-3883bp region of the gene with a lower degree of homology with other genes, and the designed primer P 1 and P 2 (SEQID NO: 1 and 2, see Table 1) Homologous genes were searched in GenBank respectively.
[0149] The search results showed that, except for rapeseed PEP gene, there was no other gene that could complement these two pairs of primers, indicating that the designed primers had strong specificity.
[0150] At the same time, will be P 1 and P 2 The target sequences of the amplified products amplified by the primers were entered into GenBank to find homologous sequences. The search results showed that, except for the PEP gene, there was no other gene sequence identical to it. Therefore, the target sequence of the amplified PCR product has high specificity.
Embodiment 3
[0152] Detection of PEP gene
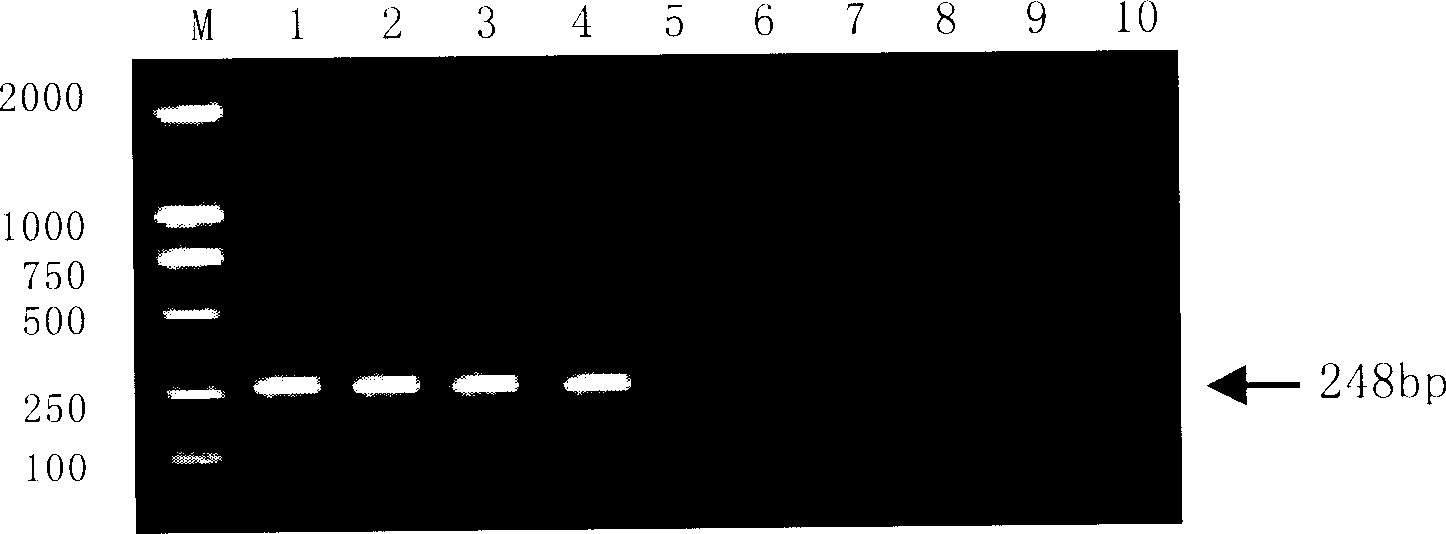
[0153] Primer P 3 ,P 4 (SEQ ID NO: 9 and 10, see Table 1) on transgenic glyphosate-resistant rapeseed, transgenic glufosinate-resistant rapeseed, non-transgenic rapeseed, rapeseed meal, soybean, corn, rice, wheat, Arabidopsis The DNA samples were tested by PCR, and a 137bp fragment could be amplified from these samples. All the above samples were extracted with Promcga Magnetic Bead Method DNA Extraction Kit.
[0154] Primer P 1 ,P 2 PCR amplification was performed on the above samples, and only 248bp amplified bands were obtained in rapeseed or rapeseed meal samples 1, 2, 3, and 4, but no PCR was found in samples 5, 6, 7, 8, and 9. Amplification ( figure 1 ). Since all DNA samples used primer P 3 ,P 4 Amplification of 137bp occurred during PCR test, primer P 3 ,P 4 It is designed based on the conserved sequence of the eukaryotic 18S RNA ribosomal gene. All eukaryotic organisms will be amplified with a 137bp PCR product, so samples 5, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com