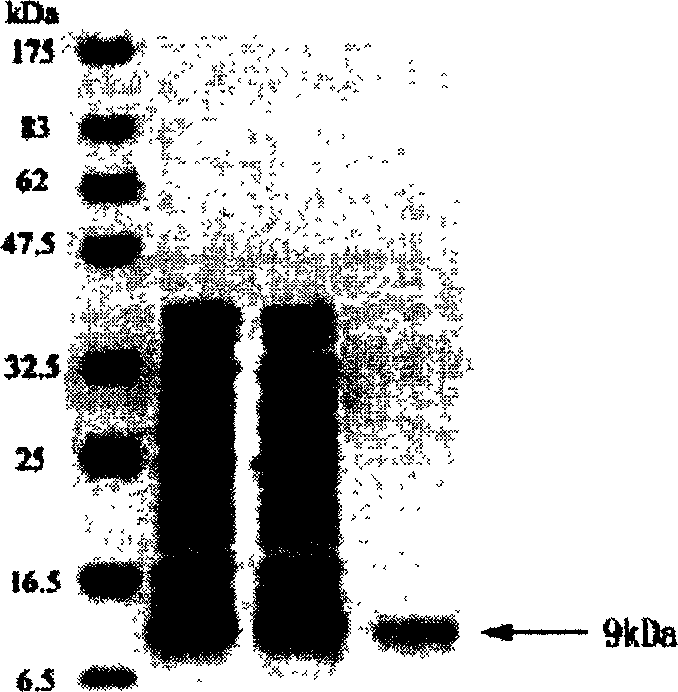Polypeptide human rudder chain dehydrogenase 9.46 and polynucleotide for encoding polypeptide
A short-chain dehydrogenase, polynucleotide technology, applied in oxidoreductase, anti-enzyme immunoglobulin, botanical equipment and methods, etc., can solve neuromuscular dysfunction, growth retardation and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0081] Example 1: Cloning of human short-chain dehydrogenase 9.46
[0082] Total RNA was extracted from human fetal brain by one-step method of guanidine isothiocyanate / phenol / chloroform. Poly(A) mRNA was isolated from total RNA using Quik mRNA Isolation Kit (product of Qiegene). 2ug poly(A) mRNA was reverse transcribed to form cDNA. Smart cDNA cloning kit (purchased from Clontech) was used to insert the cDNA fragment into the multiple cloning site of the pBSK(+) vector (product of Clontech Company), transform DH5α, and the bacteria formed a cDNA library. The sequences of the 5' and 3' ends of all clones were determined using Dye terminate cycle reaction sequencing kit (product of Perkin-Elmer) and ABI 377 automatic sequencer (Perkin-Elmer). Comparing the determined cDNA sequence with the existing public DNA sequence database (Genebank), it was found that the cDNA sequence of one of the clones, 5017C07, was a new DNA. The insert cDNA fragment contained in this clone was det...
Embodiment 2
[0083] Embodiment 2: Cloning the gene encoding human short-chain dehydrogenase 9.46 by RT-PCR method
[0084] The total RNA of fetal brain cells was used as a template, and oligo-dT was used as a primer to carry out reverse transcription reaction to synthesize cDNA. After purification with a Qiagene kit, PCR amplification was performed with the following primers:
[0085] Primer1: 5'-GCTCCTTCTGAATGCTCTGGGGCA-3'
[0086] Primer2: 5'-GCACGGCTGCGAGAAGACGAAGCT-3'
[0087] Primer1 is the forward sequence starting from the 1bp at the 5' end of 1;
[0088] Primer2 is the 3' reverse sequence in 1.
[0089] The conditions of the amplification reaction: 50mmol / L KCl, 10mmol / L Tris-Cl, (pH8.5), 1.5mmol / L MgCl in a reaction volume of 50μl 2 , 200 μmol / L dNTP, 10 pmol primer, 1 U of Taq DNA polymerase (product of Clontech Company). On a PE9600 DNA thermal cycler (Perkin-Elmer), the reaction was performed for 25 cycles under the following conditions: 94°C for 30 sec; 55°C for 30 sec; 72...
Embodiment 3
[0090] Example 3: Northern blot analysis of human short-chain dehydrogenase 9.46 gene expression:
[0091] Total RNA was extracted by a one-step method [Anal. Biochem 1987, 162, 156-159]. The method includes acid guanidine thiocyanate phenol-chloroform extraction. That is, use 4M guanidine isothiocyanate-25mM sodium citrate, 0.2M sodium acetate (DH4.0) to homogenate the tissue, add 1 times the volume of phenol and 1 / 5 volume of chloroform-isoamyl alcohol (49:1 ), mixed and centrifuged. The aqueous layer was aspirated, isopropanol (0.8 vol) was added and the mixture was centrifuged to obtain an RNA pellet. The resulting RNA pellet was washed with 70% ethanol, dried and dissolved in water. 20 µg of RNA was electrophoresed on a 1.2% agarose gel containing 20 mM 3-(N-morpholino)propanesulfonic acid (pH 7.0)-5 mM sodium acetate-1 mM EDTA-2.2M formaldehyde. Then transfer to nitrocellulose membrane. with α- 32 P dATP prepared by random primer method 32 P-labeled DNA probes. T...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com