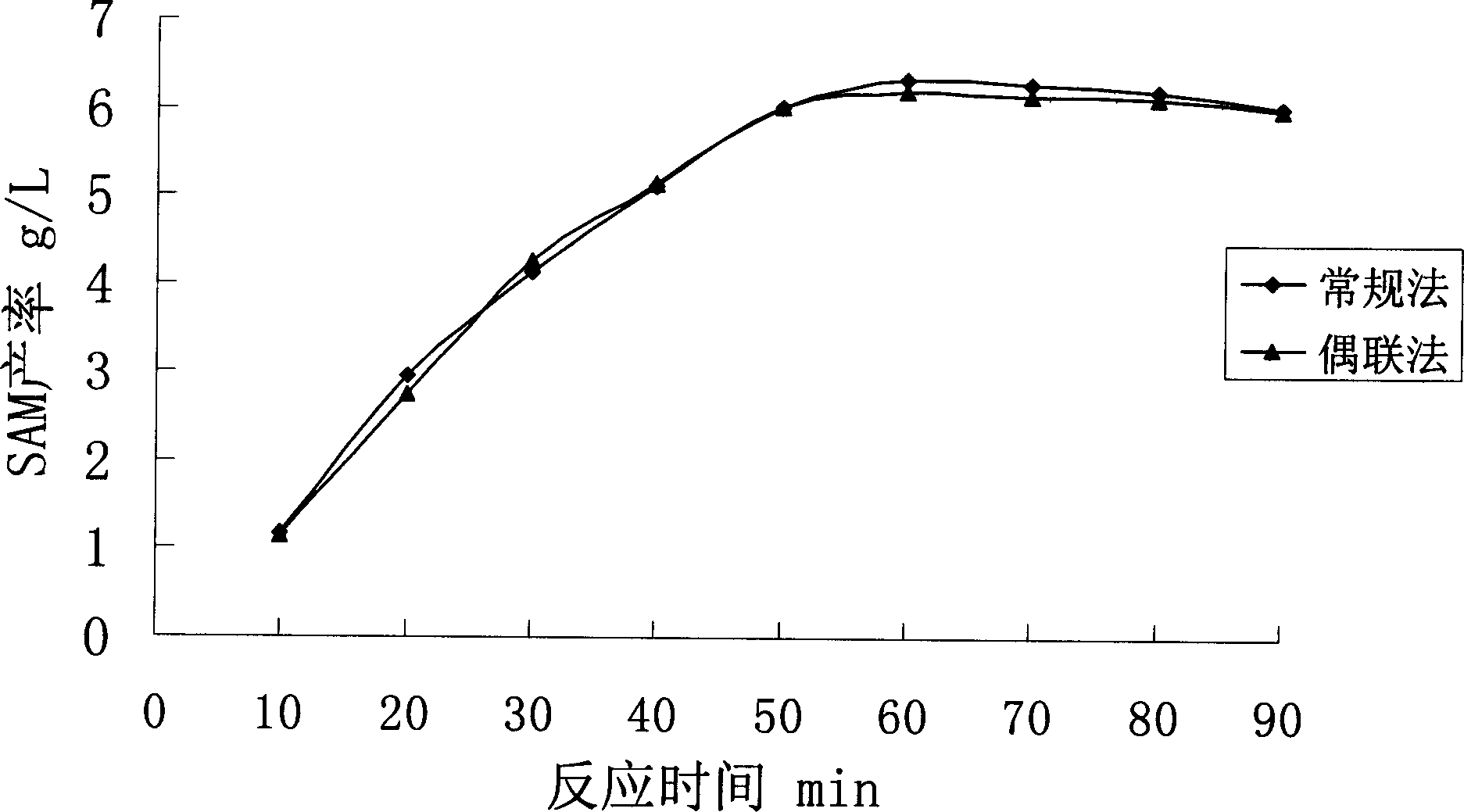
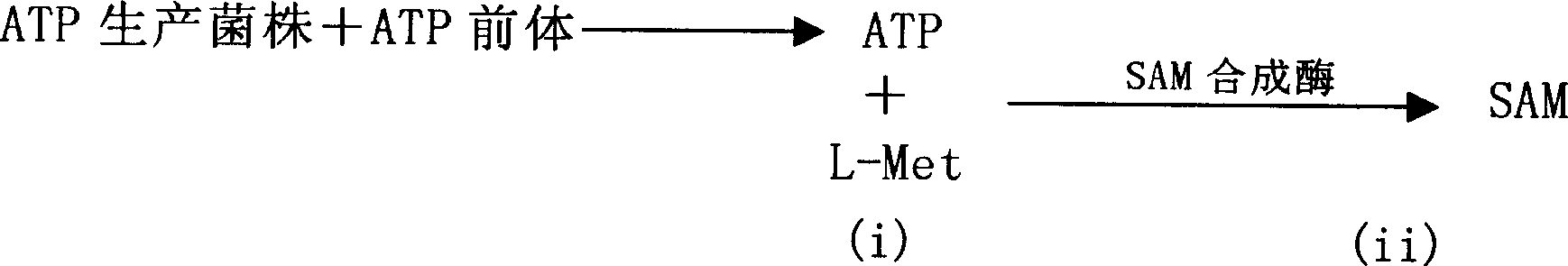Novel aden osyl methionine in vitro enzymatic transformation method
A technology of adenosylmethionine and methionine, which is applied in the field of in vitro enzymatic conversion of adenosylmethionine, which can solve the problems of difficult storage, labor and material resources, and high cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0091] ATP production by free Saccharomyces cerevisiae and SAM synthesis reaction involving free SAM synthetase
[0092] 1 material
[0093] 1.1 ATP producing strain: the strain used in this example is Saccharomyces cerevisiae CICC 1012.
[0094] 1.2 SAM synthetase producing strain: The SAM synthetase producing strain used in this example has a strain preservation number of CGMCC, NO.0648, provided by the Institute of Biochemistry and Cell Biology, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, and its construction process can be found in CN1357630A.
[0095] 1.3 Lysis buffer:
[0096] Phosphate buffer 50mM / L, glycerin 5%, EDTA 1mM / L, PMSF 0.5mM / L
[0097] 1.4 Bioreactor: FUS-15L fermenter, produced by Shanghai Guoqiang Biochemical Engineering Equipment Co., Ltd.
[0098] 1.5 Medium: YPD medium: yeast extract 10g / L, peptone 20g / L, glucose 20g / L.
[0099] 1.6 Mixed reaction solution
[0100] L-methionine 10mM / L, glutathione 16mM / L, dipotassium hy...
Embodiment 2
[0112] ATP production by immobilized Saccharomyces cerevisiae and SAM synthesis reaction involving immobilized SAM synthetase
[0113] 1. Materials
[0114] 1.1 ATP producing strain: the strain used in this example is Saccharomyces cerevisiae CICC 1202.
[0115] 1.2 The production strain of SAM synthetase, yeast lysate buffer, bioreactor or shake flask, and YPD medium required for yeast growth are all the same as in Example 1.
[0116] 1.3 The reaction solution for coupling immobilized yeast with immobilized SAM synthetase contains:
[0117] L-methionine 10mM / L, glutathione 16mM / L, potassium chloride 200mM / L, dipotassium hydrogen phosphate 200mM / L, Ado 40mM / L, glucose 150mM / L, magnesium sulfate 20mM / L, Tris - HCl buffer 10 mM / L; reaction pH 7.0.
[0118] 2 methods
[0119] 2.1 The preparation of SAM synthetase is the same as in Example 1.
[0120] 2.2 Immobilization of SAM synthetase
[0121] Dilute the initially obtained crude enzyme solution with a pH 6.8 phosphate buf...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com