Gene for coding penicillium chrysogenum phenylacetic acid hydroxylase and its use
A technology for the activity and encoding of phenylacetic acid hydroxylase, which is applied in the fields of enzymes, biochemical equipment and methods, and the introduction of foreign genetic material using vectors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] [Example 1]: pahB gene cloning
[0033] Cultivate Penicillium chrysogenum Wis54-1255 (ATCC28089) in YPD (1% yeast extract, 2% peptone, 2% glucose) medium for 24h, filter and collect mycelium, take 0.1g of wet mycelium liquid N 2 After grinding, add 1ml Trizol reagent (Invitrogen, 15596-026) to extract the total RNA of Penicillium chrysogenum, dissolve the obtained total RNA of Penicillium chrysogenum in DEPC water, and store it at -70°C for future use.
[0034] To remove possible DNA contamination, take 5 μg of total RNA from Penicillium chrysogenum, add 5 μl RQ1 RNase-Free DNase (Promega, M6101), 10 μl RQ1 RNase-FreeDNase reaction buffer (10×), and add 5 μl RQ1 RNase-Free DNase reaction buffer (10×) to the 100 μl reaction system. Incubate at ℃ for 30 minutes, extract with phenol: chloroform, precipitate with ethanol, dissolve in 10 μl DEPC water. Take 5 μl of the DNA-depleted RNA sample and perform reverse transcription with random primers (RandomHexamers, Promega, C1...
Embodiment 2
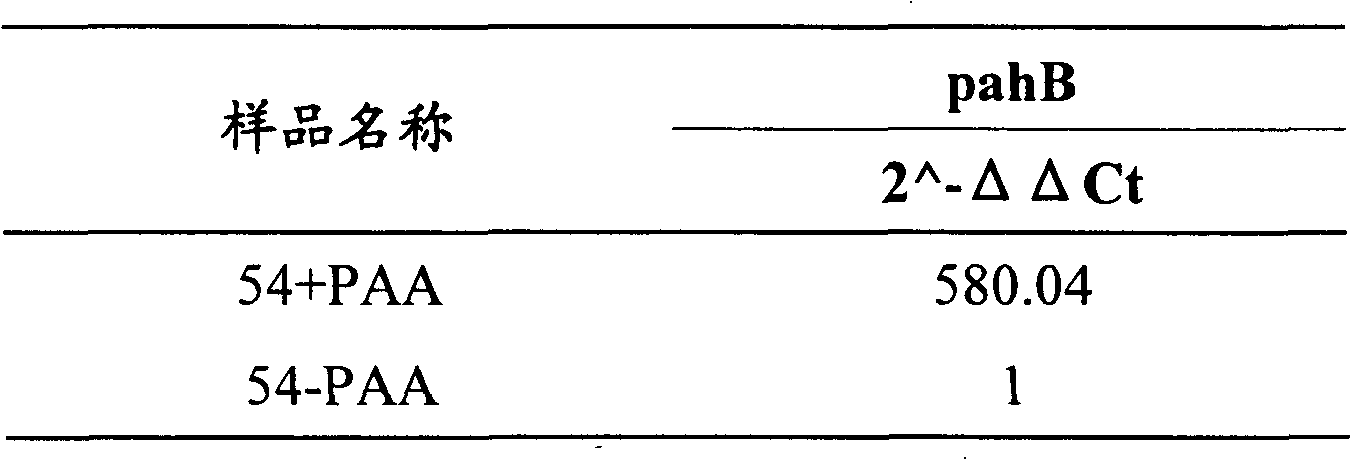
[0038] [Example 2]: The expression of pahB gene is induced by phenylacetic acid
[0039] Penicillium chrysogenum Wis54-1255 (ATCC28089) was pressed 2×10 6 Conidia / ml was inoculated into the seed medium and cultured at 26°C for 24 hours. The formulation of the seed medium was (g / l): glucose, 30; lactose, 10; cottonseed meal, 10; yeast powder, 10; corn pulp, 10; (NH 4 ) 2 SO 4 , 2; KH 2 PO 4 , 1; CaCO 3 , 5; pH5.8. The seed culture was then inoculated into the fermentation medium at a 5% inoculum size and cultured at 26°C. The fermentation medium is (g / l): lactose, 120; cottonseed meal, 30; corn steep liquor, 20; (NH 4 ) 2 SO 4 , 5; KH 2 PO 4 ,1;K 2 SO 4 , 5; CaCO 3 , 10; pH 6.5.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com