Gene chip for inspecting and grouping SARS coronavirus, its method and reagent kit
A coronavirus and gene chip technology, applied in the field of detection, can solve problems such as inability to type and predict epidemics, and achieve high accuracy, strong repeatability, and easy operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example one Preparation of oligonucleotide probes
[0033] 1. Sequence alignment: Use the multiple sequence alignment software clustalx to compare the sequences of the s genes of 38 different SARS coronaviruses, and obtain 27 mutation sites in the s genes. According to the number of mutation sites, they can be divided into 4 types: prototype (no mutation), low pathogenic type (mutation at 2-7 sites), high pathogenic type (occurring at 17-25 sites) Mutation) and epidemic type (27 sites have mutations).
[0034]2. Design probe: Use Primer5 biology software to design 106 oligonuclei for typing based on 25 mutation sites (the other two sites did not cause amino acid changes after mutations, so they are not tested) In addition, two probes were designed to detect SARS coronavirus in the conserved region of the s gene. The 108 oligonucleotide probes mentioned above are shown in Table 1.
[0035] Table 1. Oligonucleotide probes for SARS coronavirus typing and detection
[0036] ...
Embodiment 2
[0041] Example two Preparation of primers for amplification of s gene
[0042] 1. Primer design: Use Primer 5.0 and other bioinformatics software to analyze the genome of SARS coronavirus, and design 4 pairs of primers to amplify the s gene in the conserved region of the s gene. The primer sequence is shown in Table 2.
[0043] Table 2. Primers used to amplify the s gene
[0044] SEQ ID
[0045] 2. Primer synthesis: send the selected primer sequence to the primer synthesis company for entrusted synthesis (synthesized by Aoke Company) for use.
Embodiment 3
[0046] Example three Chip preparation
[0047] 1. Preparation of probe solution: Dissolve the probe prepared in Example 2 in a 50% DMOS solution so that the final concentration of the probe reaches 1 μg / μl.
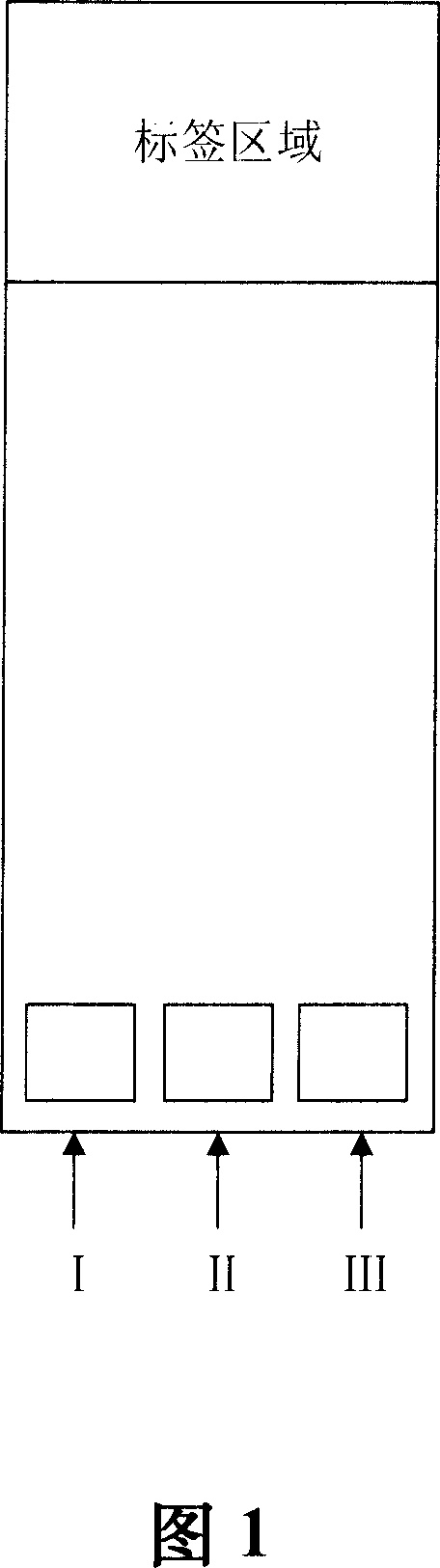
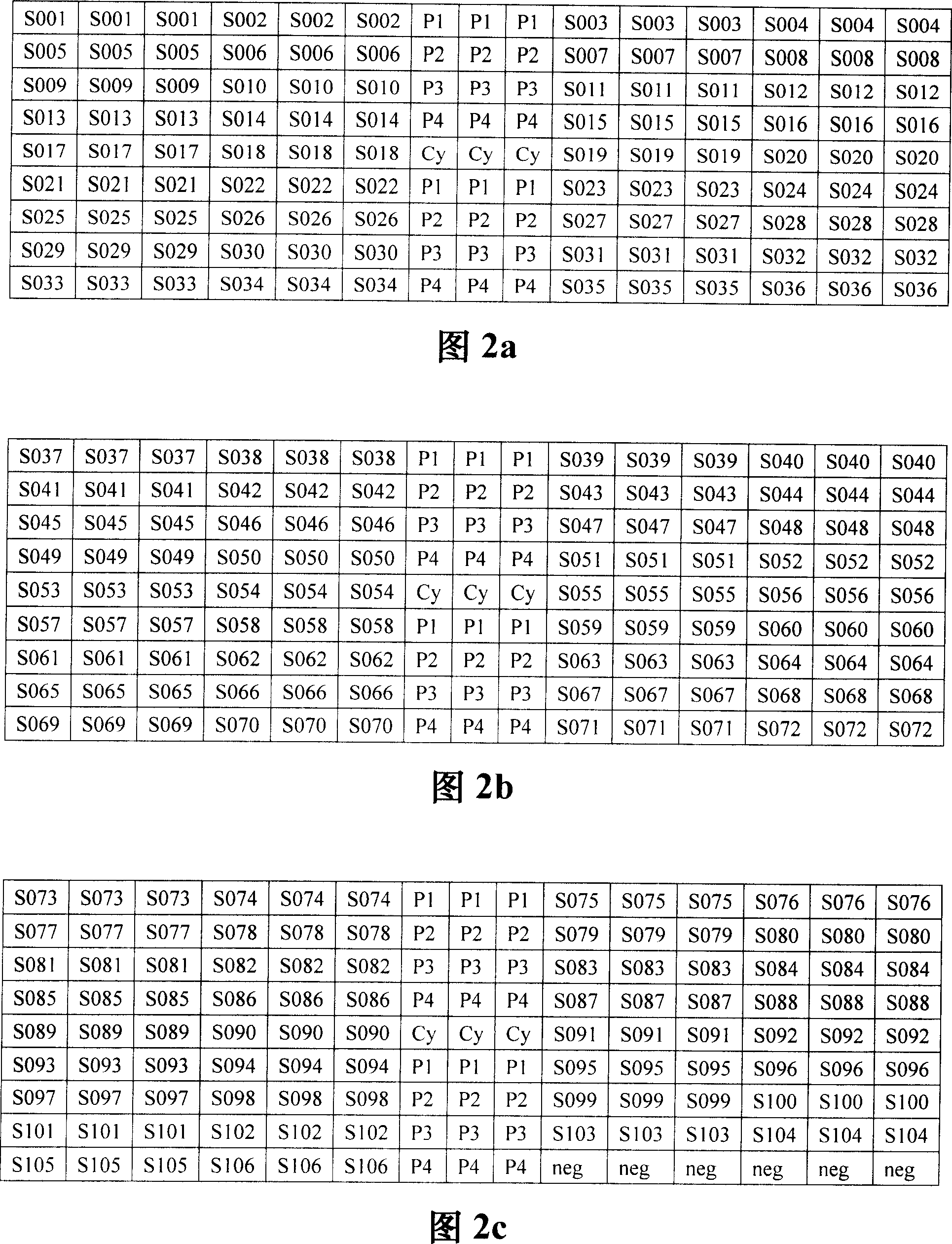
[0048] 2. Chip spotting: Take 57.5mm×25.5mm×1mm (length×width×height) aldehyde-based glass slide as the solid phase carrier, on which there are three spotting areas I, II, and III (as shown in the figure) 1). Use the automatic biochip spotting instrument (Spotarray 72) to spot samples in the three spotting areas with the 9 (row) × 15 (column) probe arrangements shown in Figures 2a, 2b, and 2c to make a gene chip. As shown in Figures 2a, 2b and 2c, Cy represents oligonucleotides labeled with fluorescein Cy3. These points are used to determine the start and end positions of the array during the scanning process; P1-P2 represent the positive control used to detect SARS coronavirus Probe; neg stands for negative control probe; S001-S106 stands for specific probes used to detect...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com