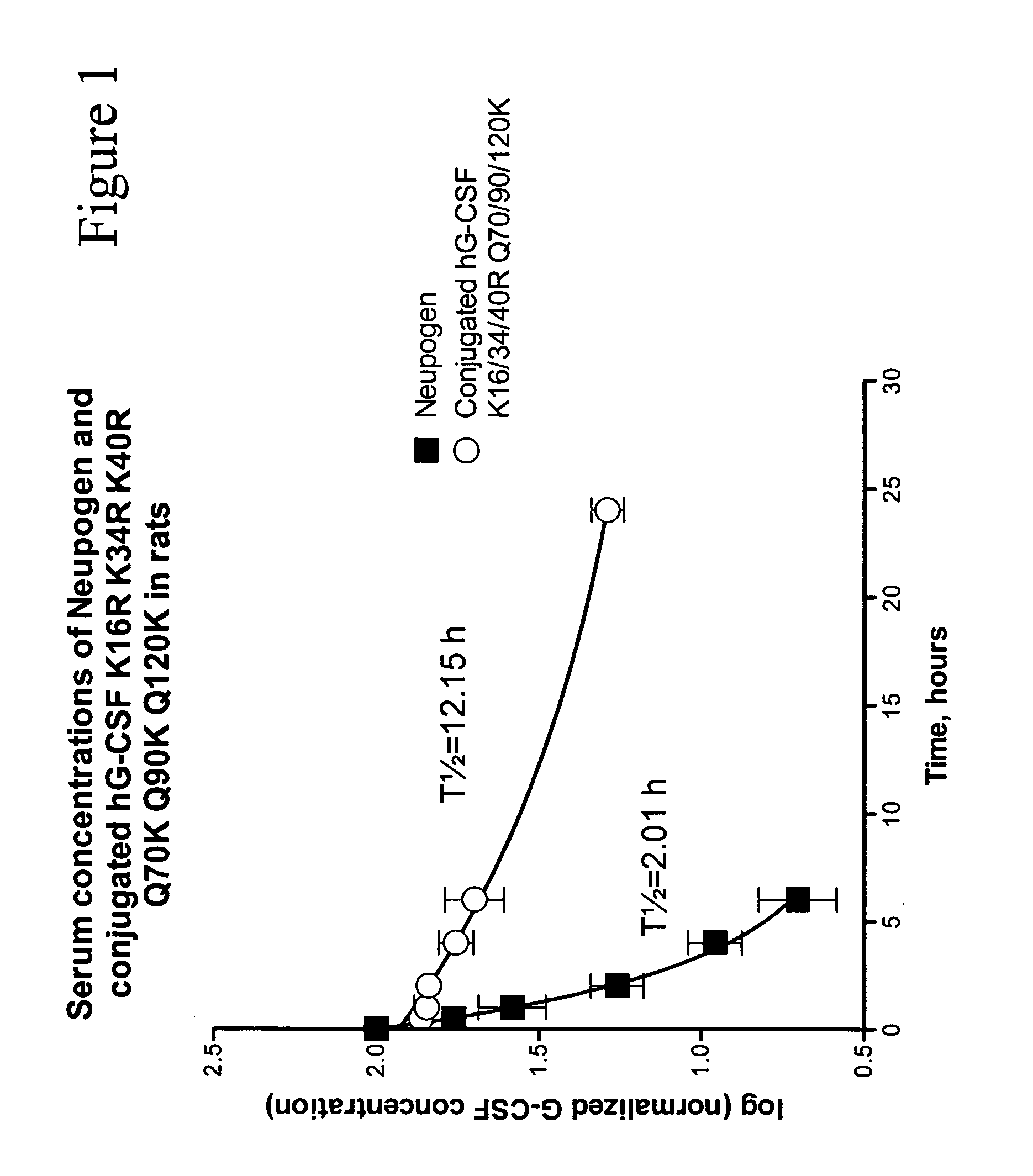
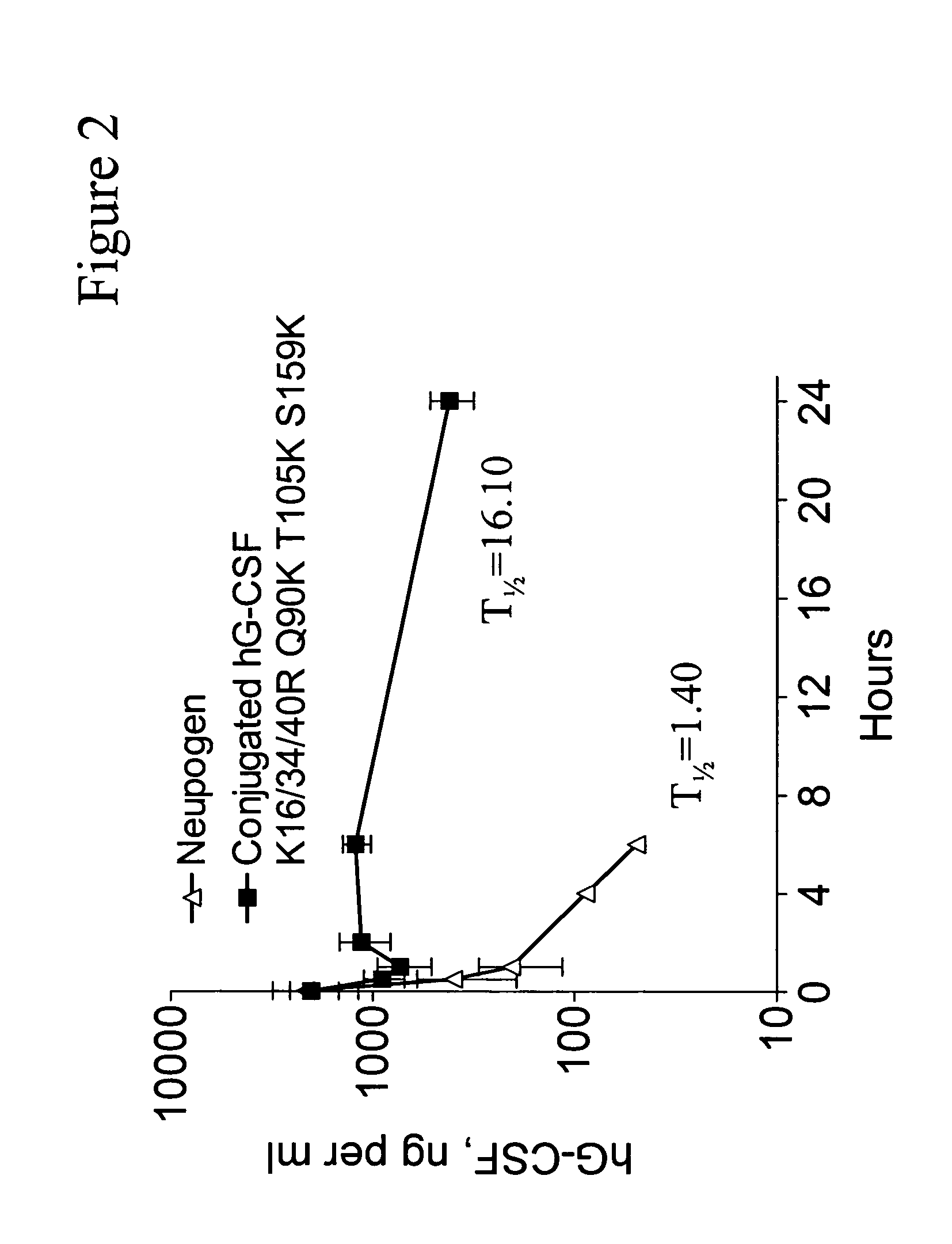
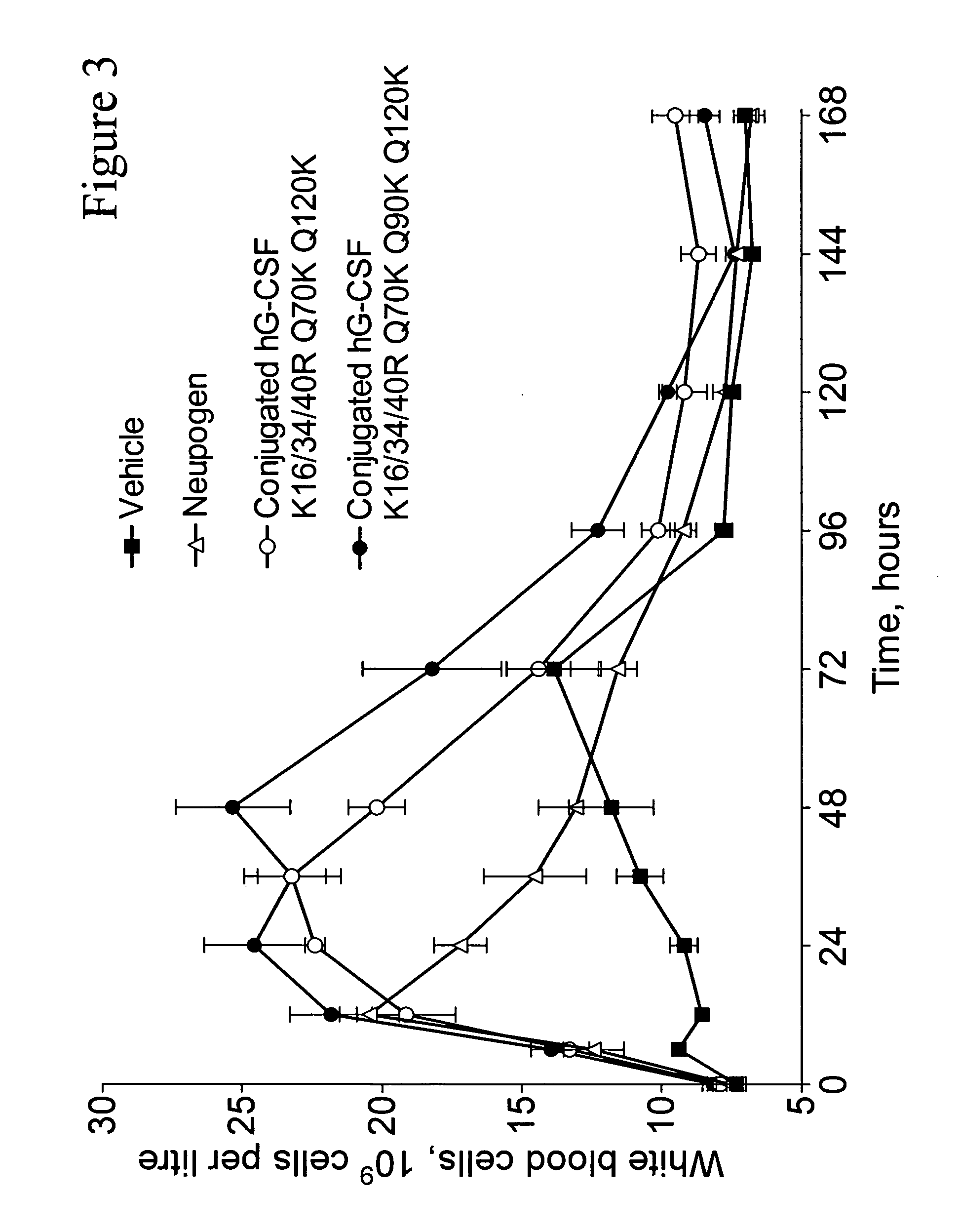
G-CSF conjugates
a granulocyte colony-stimulating factor and conjugate technology, applied in the direction of peptides/protein ingredients, animal/human proteins, peptides, etc., can solve the problems of dose-dependent bone pain, relative minor infections can be serious and even life-threatening, and patients become neutropenic after chemotherapy, etc., to reduce in vitro bioactivity, improve properties, and rapid neutrophil recovery
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction and Cloning of Synthetic Genes Encoding hG-CSF
[0269] The following DNA fragments were synthesized following the general procedure described by Stemmer et al. (1995), Gene 164, pp. 49-53:
[0270] Fragment 1, consisting of a Bam HI digestion site, a sequence encoding the YAP3 signal peptide (WO 98 / 32867), a sequence encoding the TA57 leader sequence (WO 98 / 32867), a sequence encoding a KEX2 protease recognition site (AAAAGA), a sequence encoding hG-CSF with its codon usage optimized for expression in E. coli, (SEQ ID NO:2) and a Xba I digestion site.
[0271] Fragment 2, consisting of a Bam HI digestion site, a sequence encoding the YAP3 signal peptide (WO 98 / 32867), a sequence encoding the TA57 leader sequence (WO 98 / 32867), a sequence encoding a histidine tag (SEQ ID NO:5), a sequence encoding a KEX2 protease recognition site (AAAAGA), a sequence encoding hG-CSF with its codon usage optimized for expression in E. coli, (SEQ ID NO:2) and a Xba I digestion site.
[0272] Fra...
example 2
Expression of hG-CSF in S. cerevisiae and E. coli
[0277] Transformation of Saccharomyces cerevisiae YNG318 (available from the American Type Culture Collection, VA, USA as ATCC 208973) with either plasmid pG-CSFcerevisiae or pHISG-CSFcerevisiae, isolation of transformants containing either of the two plasmids, and subsequent extracellular expression of hG-CSF without and with the HIS tag, respectively, was performed using standard techniques described in the literature. Transformation of E. coli BL21 (DE3) (Novagen, Cat. No. 69387-3) with pG-CSFcoli, isolation of transformants containing the plasmid and subsequent expression of hG-CSF in the supernatant and in the periplasm of the cell was performed as described in the pET System Manual (8th edition) from Novagen.
[0278] Expression of hG-CSF by S. cerevisiae and E. coli was verified by Western Blot analysis using the ImmunoPure Ultra-Sensitive ABC Rabbit IgG Staining kit (Pierce) and a polyclonal antibody against hG-CSF (Pepro Tech...
example 3
Generation of a Stable CHO-K1 G-CSF Producer
[0280] The day before transfection the CHO K1 cell line (ATCC #CCl-61) is seeded in a T-25 flask in 5 ml DMEM / F-12 medium (Gibco # 31330-038) supplemented with 10% FBS and penicillin / streptomycin. The following day (at nearly 100% confluency) the transfection is prepared: 90 μl DMEM medium without supplements is aliquoted into a 14 ml polypropylene tube (Corning). 10 μl Fugene 6 (Roche) is added directly into the medium and incubated for 5 min at room temperature. In the meantime 5 μg plasmid pG-CSFCHO is aliquoted into another 14 ml polypropylene tube. After incubation the Fugene 6 mix is added directly to the DNA solution and incubated for 15 min at room temperature. After incubation the whole volume is added drop-wise to the cell medium.
[0281] The next day the medium is exchanged with fresh medium containing 360 μg / ml hygromycin (Gibco). Every day hereafter the selection medium is renewed until the primary transfection pool has reach...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com