Disintegrin homologs
a technology of disintegrin and homologs, applied in the field of disintegrin homologs, can solve problems such as affecting the binding of fibrinogen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
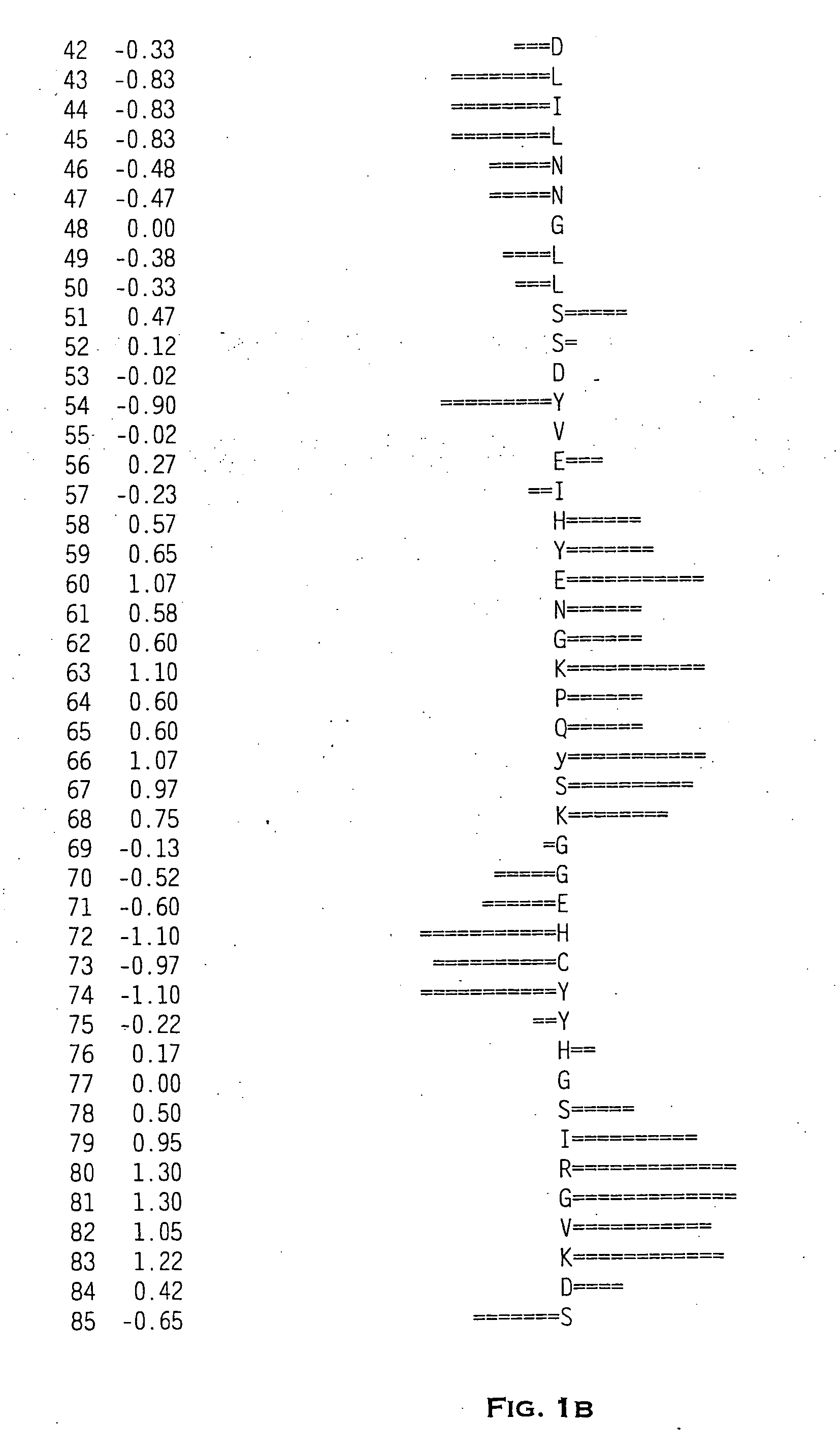
Image
Examples
example 1
Extension of EST Sequence
[0196] The novel zdint1 polypeptide-encoding polynucleotides of the present invention were initially identified by querying an EST database. This query identified an expressed sequence tag (EST) to nucleotide 1097 to nucleotide 1415 of SEQ ID NO: 1. A cDNA clone, corresponding to this EST was obtained and the deduced amino acid sequence was determined to be incomplete. Primers ZC17,991 (SEQ ID NO:4) and ZC17,992 (SEQ ID NO:5) were used to screen an arrayed fetal brain cDNA plasmid library to identify clones of zdint1. Thermocycler conditions were as follows: one cycle at 94° C. for 1 minute 30 seconds; followed by thirty cycles at 94° C. for 10 seconds, 64° C. for 20 seconds, 72° C. for 30 seconds, followed by one cycle at 72° C. for 5 minutes, followed by a 4° C. hold. A sample of the reaction contents was electrophoresed on a 4% agarose gel to identify positive pools. These pools were screened by polymerase chain reaction with ZC17,992 (SEQ ID NO:5) and t...
example 2
[0197] Analysis of tissue distribution was performed by the Northern blotting technique using Human Multiple Tissue and Master Dot Blots from Clontech (Palo Alto, Calif.), and a human vascular tissue blot prepared in-house. The human vascular blot was prepared from the following cell lines: Human Umbilical Vein Endothelial Cells (Cascade Biologics, Inc., Portland, Oreg.), Human Pulmonary Artery Endothelial Cells (Cascade Biologics, Inc., Portland, Oreg.), Human Aortic Endothelail Cells, (Cascade Biologics, Inc., Portland, Oreg.), Aortic Smooth Muscle Cells (Clonetics, San Diego, Calif.), Human Intestinal Smooth Muscle Cells (American Type Culture Collectio, Manasas, Va.), Normal Human Lung Fibroblast, Clonetics, San Diego, Calif.) and Normal Human Dermal Fibroblast-Neonatal, Clonetics, San Diego, Calif.). Messenger RNA was extracted and blots prepared by methods known in the art. The probe was obtained by restriction digest of the original cDNA clone with a restr...
example 3
Protein Purification
[0198] Purification conditions for zdint1 with N- and C-terminal EE tags:
[0199]E. coli, Pichia, CHO and BHK cells are transfected with expression vectors containing the DNA sequence of SEQ ID NO:1, or a portion thereof, operably linked to a polynucleotide encoding a Glu-Glu tag. Zdint1 protein is expressed in conditioned media of E. coli, Pichia methanolica, and or chinese hamster ovary (CHO) and baby hamster kidney (BHK) cells. For zdint1 expressed in E. coli and Pichia, the media is not concentrated prior to purification. Unless otherwise noted, all operations are carried out at 4° C. A total of 25 liters of conditioned medium from BHK cells is sequentially sterile filtered through a 4 inch, 0.2 mM Millipore (Bedford, Mass.) OptiCap capsule filter and a 0.2 mM Gelman (Ann Arbor, Mich.) Supercap 50. The material is then concentrated to about 1.3 liters using a Millipore ProFlux A30 tangential flow concentrator fitted with a 3000 kDa cutoff Amicon (Bedford, Mas...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Length | aaaaa | aaaaa |
| Interaction | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com