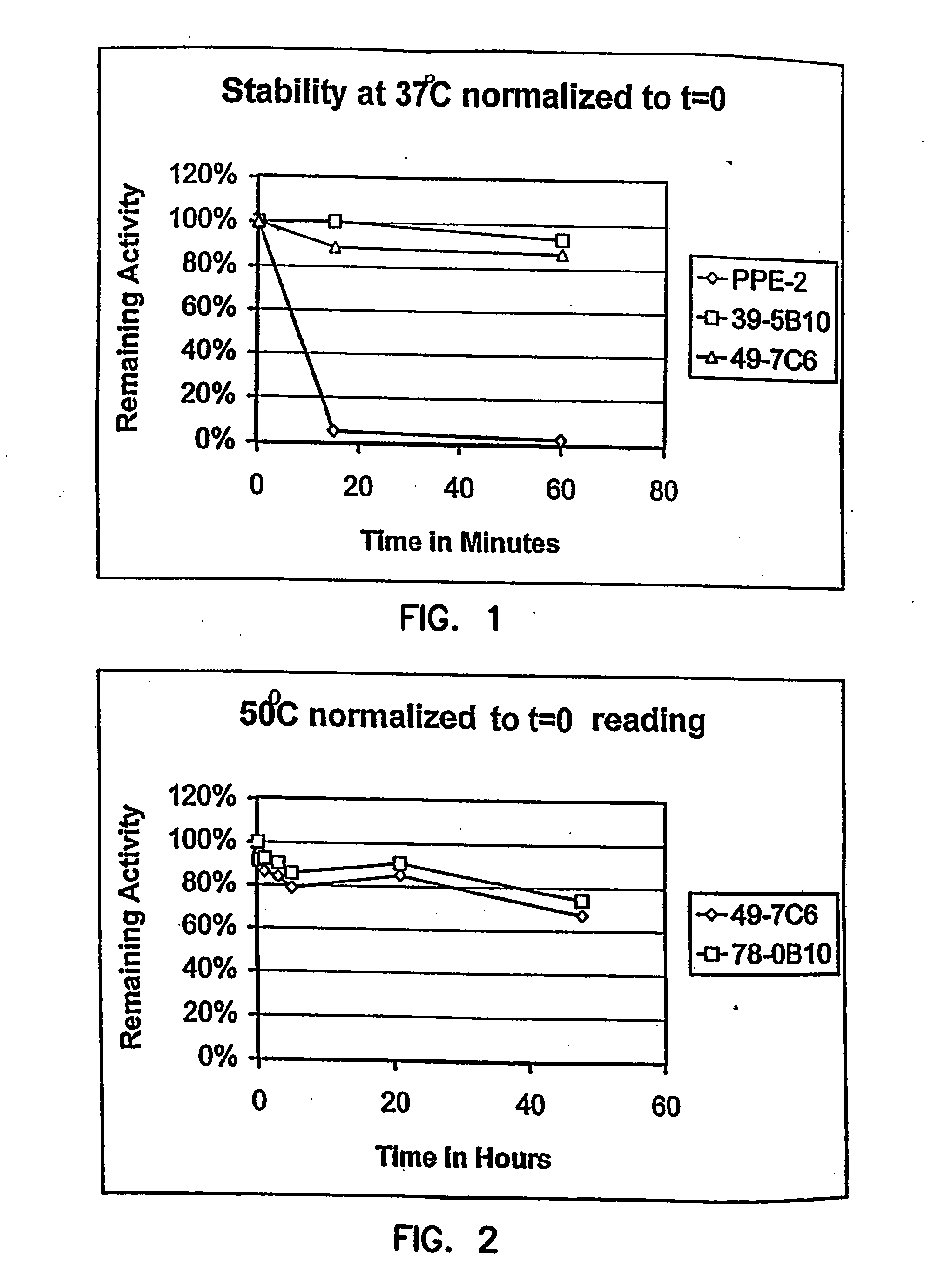
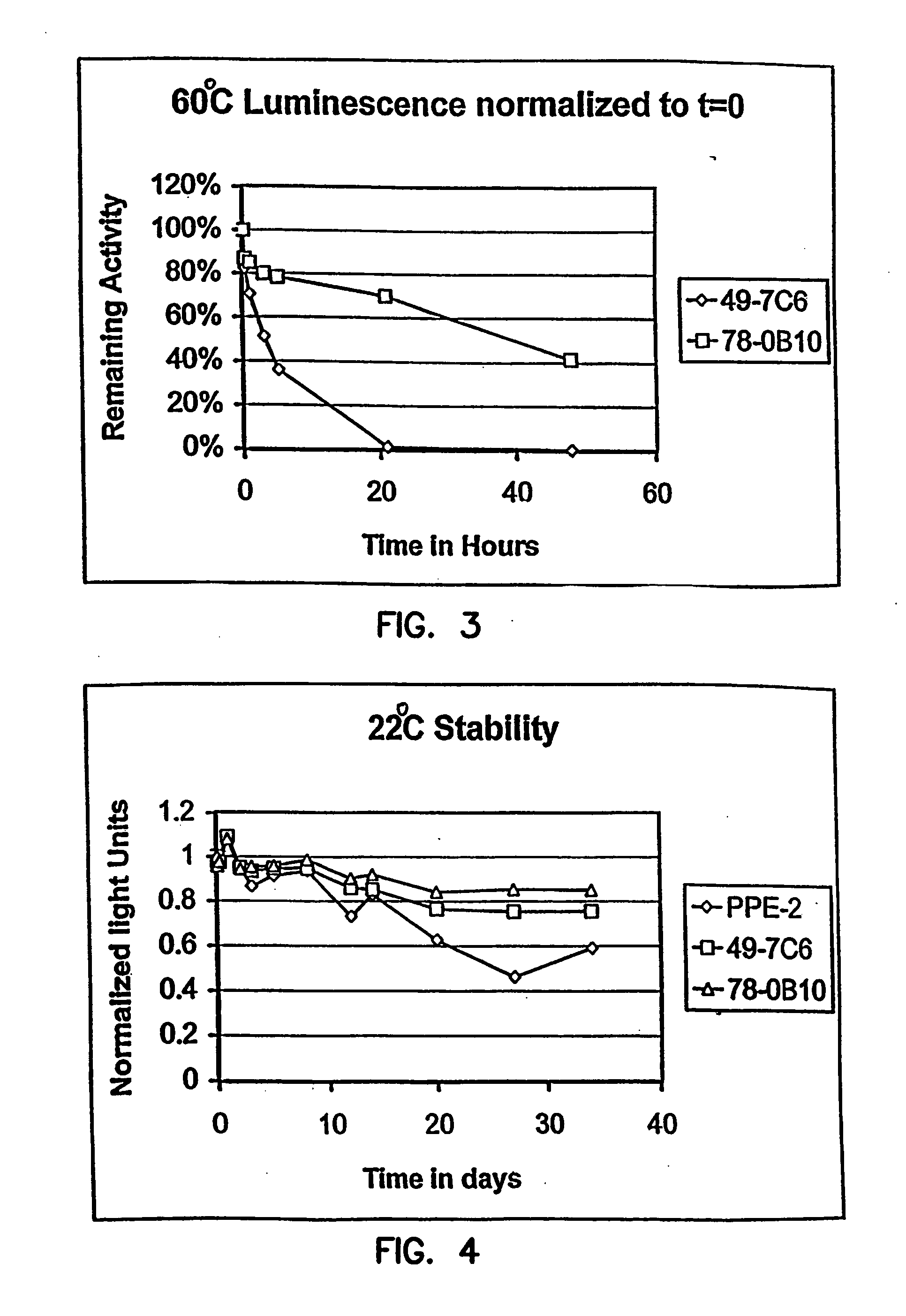
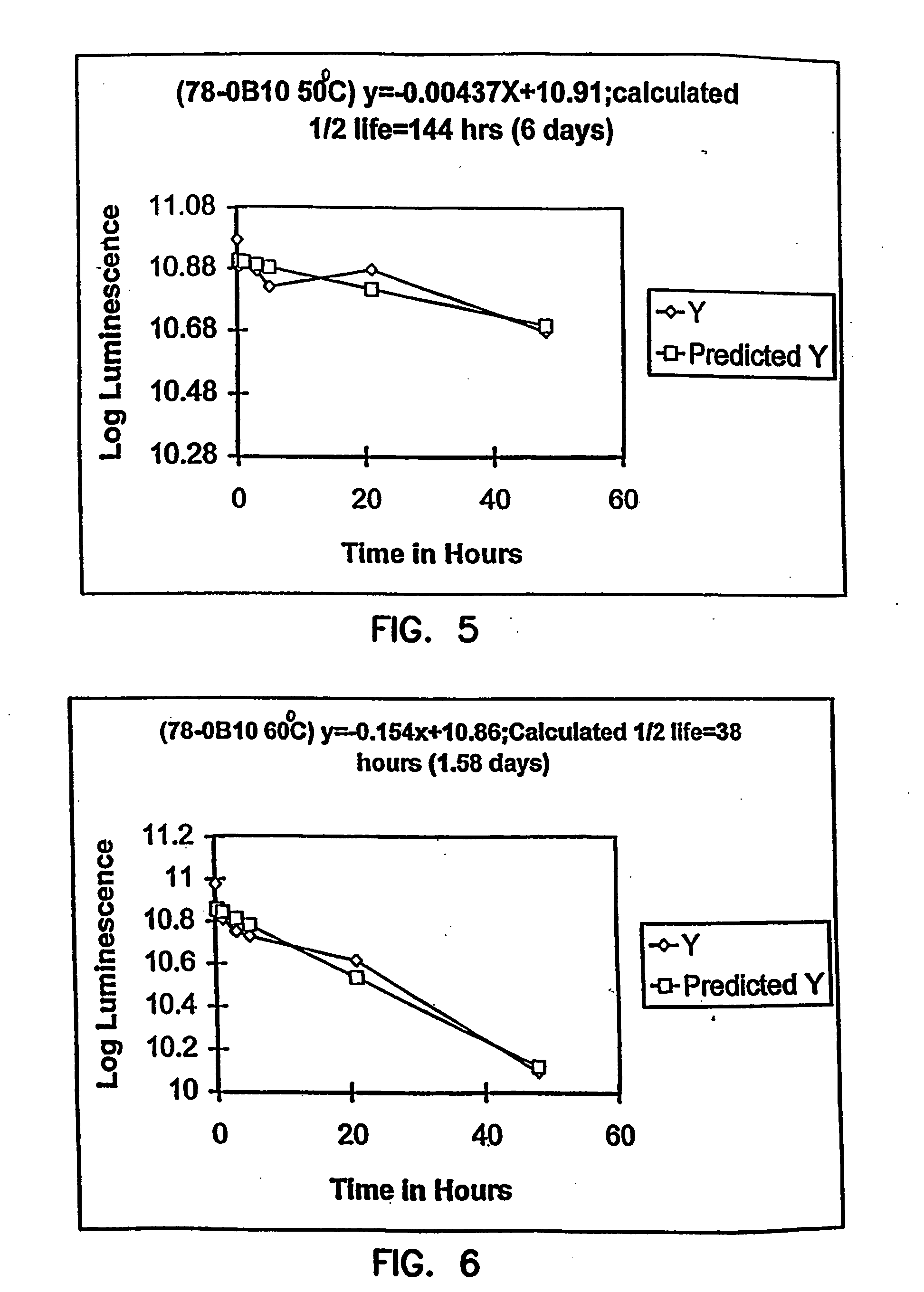
Thermostable luciferases and methods of production
a technology of luciferases and luciferases, which is applied in the field of luciferase enzymes with mutants, can solve the problems of limited general utility of beetle luciferases, and the stability of beetle luciferases having amino acid sequences encoded by cdna sequences cloned from luminous beetles, so as to achieve enhanced luminescence intensity, less than 5% luminescence activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Producing Thermostable Luciferases of the Present Invention
Mutagenesis Method.
[0131] An illustrative mutagenesis strategy is as follows: From the “best” wild-type luciferase clone, that is a clone with increased thermostability and not appreciably diminished values for other parameters, random mutagenesis was performed by three variations of error-prone PCR. From each cycle of random mutagenesis, 118 of the best clones were selected. DNA was prepared from these clones yielding a total of 54 clones. These clones represent new genetic diversity.
[0132] These 54 clones were combined and recombination mutagenesis was performed. The 18 best clones from this population were selected.
[0133] These 18 clones were combined with the 18 clones of the previous population and recombination mutagenesis was performed. From this screening, a new luciferase population of 18 clones was selected representing 6 groups of functional properties.
[0134] In this screening the new mutations of the select...
example 2
Organize Data into SOL Database
[0277] Each file created by a luminometer (96 well, Anthos, Austria) represents the data from one microplate. These files are stored in the computer controlling the luminometer, and connected to the database computer by a network link. From each microplate of samples, nine microplates are read by the luminometer (the original microplate for optical density and eight daughter microplates for luminescence).
[0278] Ninety files are created in total; each containing data sets for 96 samples. Each data set contains the sample number, time of each measurement relative to the first measurement of the plate, luminometer reading, and background corrected luminometer reading. Other file header information is also given. The time that each microplate is read is also needed for analysis. This can be obtained from the robot log or the file creation time. A naming convention for the files is used by the robot during file creation that can be recognized by...
example 3
Preparation of Novel Luciferases
[0393] The gene shown in FIG. 45 contains a single base pair mutation which encodes an amino acid substitution at position 249, T to M. This clone has a spectral maximum of 552 nm which is yellow shifted from the sequence of Luc. This mutant was selected as an original template because it produces about 5 times brighter luminosity in vivo which allowed for more efficient screening.
C-Terminus Mutagenesis
[0394] To eliminate the peroxisome targeting signal (SKL), the L was mutated to a STOP codon and the 3 codons immediately upstream were randomized according to the oligonucleotide mutagenesis procedure described herein. The mutagenic oligonucleotide designed to accomplish this also introduces a unique SpeI site to allow mutant identification without sequencing. The mutants were screened in vivo and 13 colonies picked, 12 of which contained the SpeI site.
N-Terminus Mutagenesis
[0395] To test if expression could be improved, the 3 codons immediately...
PUM
| Property | Measurement | Unit |
|---|---|---|
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com