Method for down-regulating gene expression in fungi
a gene expression and fungus technology, applied in the field of down regulation of gene expression in fungi, can solve the problems of only achieving rnai, cells are useful for experimental studies within the laboratory, and are clearly not suitable for many potential practical applications
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning of Target Genes
[0226] The methods for dsRNA mediated gene silencing in fungi as described herein are applicable to combat plant pathogenic fungi with dsRNA corresponding to a plant pathogen target gene. Suitable plant pathogen target genes were identified and isolated from the fungus as follows.
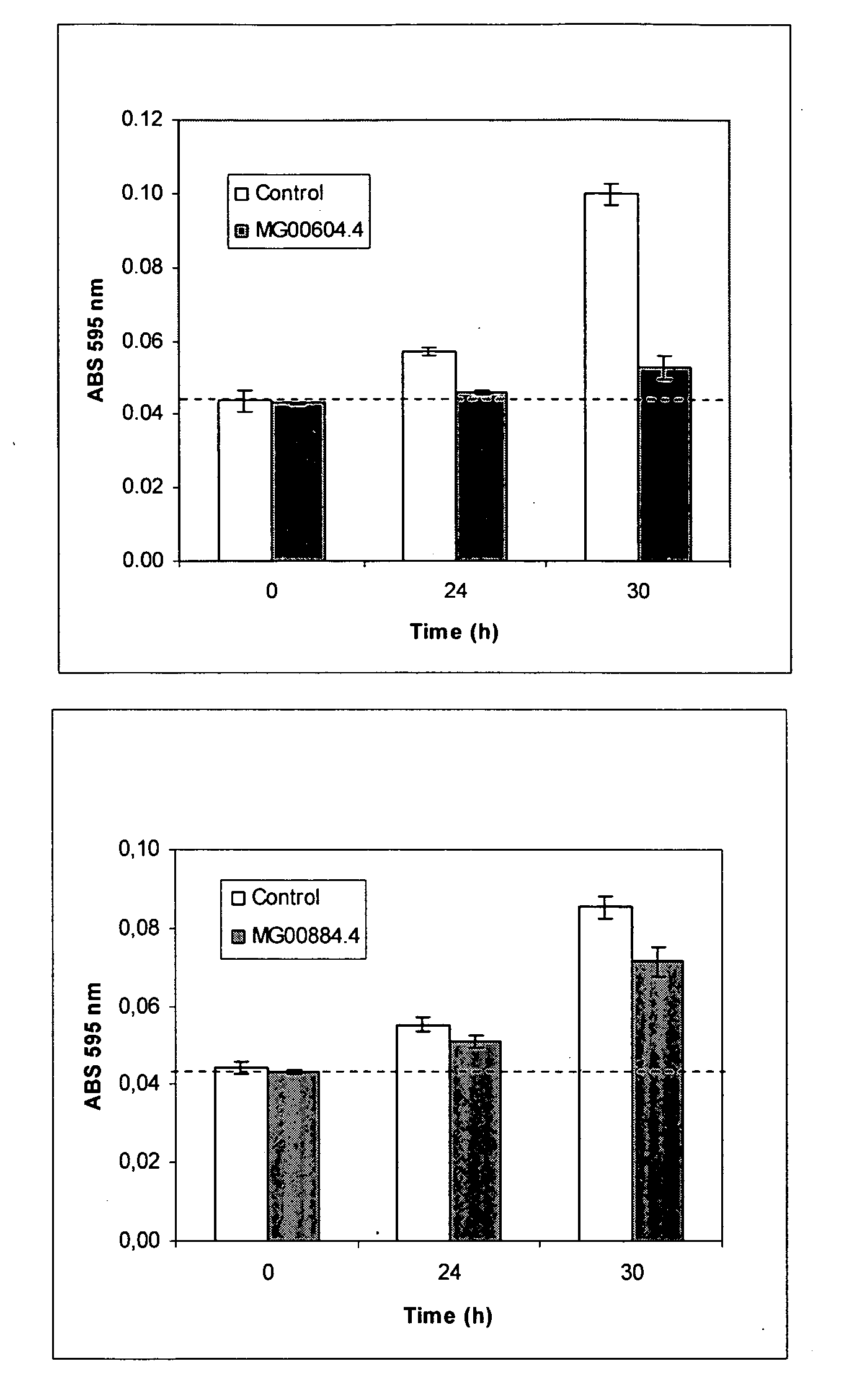
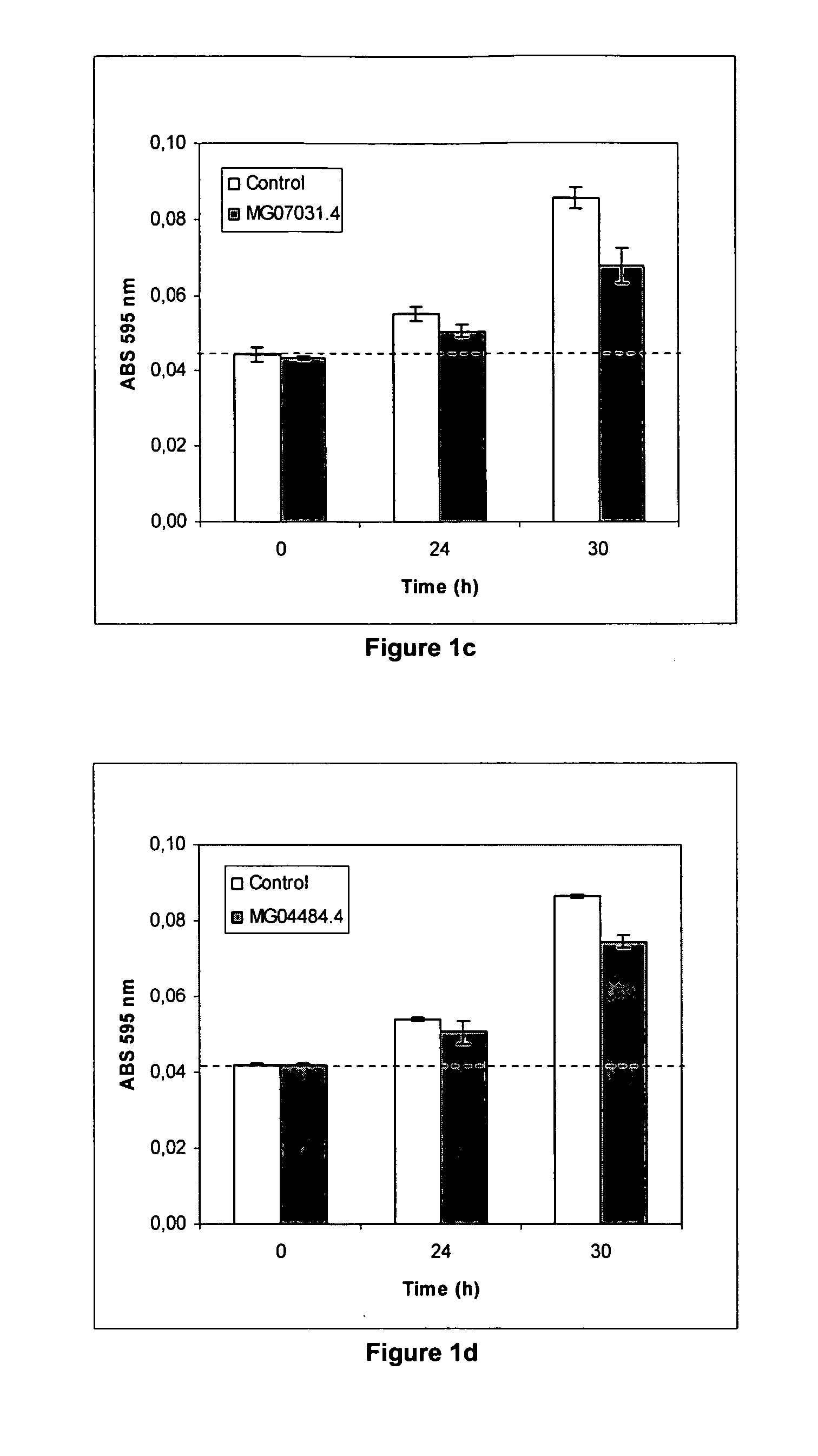
[0227] Total RNA was prepared from the fungus Magnaporthe grisea using the RNeasy Mini kit for plants (QIAGEN Cat. No. 74904), from which cDNA was prepared using the Superscript™ Double-Stranded cDNA synthesis kit (Invitrogen). To isolate the coding sequence of the Magnaporthe grisea target genes MG00884.4, MG04484.4, MG07472.4, MG06292.4, MG03668.4, MG05169.4 and MG03872.4, PCRs were performed on the Magnaporthe grisea cDNA or on genomic DNA as a template.
[0228] The PCR conditions and primers for the target gene are outlined in the tables. Each PCR was performed in duplicate. The resulting two independent PCR products per target gene, were analysed on agarose gel, isolated and clo...
example 2
Optimization of Fragment Length
[0243] The beta-tubulin was used to optimize the size of fragments that give color effect in the color assay (Example 4.3). 200, 400 and 600 bp fragments from the 3′ end of the coding region were tested in the color assay and a 400 bp, E7F7 (FIG. 6), was found to be optimal for causing a lighter color. Subsequently, for all the targets, 400 bp dsRNA was used in the color assay. Spores from two Magnaporthe grisea isolates, R67 (FIG. 5) and B157 (FIGS. 7 & 8) gave a similar color phenotype in this assay.
example 3
Selection of Target Nucleotide Sequences of the Target Genes of Magnaporthe grisea for RNAi Mediated Gene Silencing
[0244] Fragments of the target genes herein described were selected for use in further RNAi experiments both in vitro as described in example 4 or in vivo as described in examples 5, 6 and 7. These fragments are listed in Table 5. (PY maybe A person skilled in the art will recognize that other fragments of various lengths may be identified in the Magnaporthe grisea sequences, and that the present invention also extends to these fragments and the use thereof in RNAi mediated silencing of fungal genes.
[0245] These fragments or target nucleotide sequences of fungal target genes were used to produce dsRNA in vitro as is described in example 4, or were cloned in a hairpin construct to produce dsRNA in a plant cell (see example 5).
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com