Monoclonal antibodies specific for anthrax spores and peptides derived from the antibodies thereof
a technology of anthrax spores and monoclonal antibodies, which is applied in the field of monoclonal antibodies specific for anthrax spores and peptides derived from the antibodies thereof, can solve the problems of lack of accuracy and limited sensitivity, complex methods, and high cost of systems, and achieves low cost and high sensitivity. the effect of reducing the risk of infection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
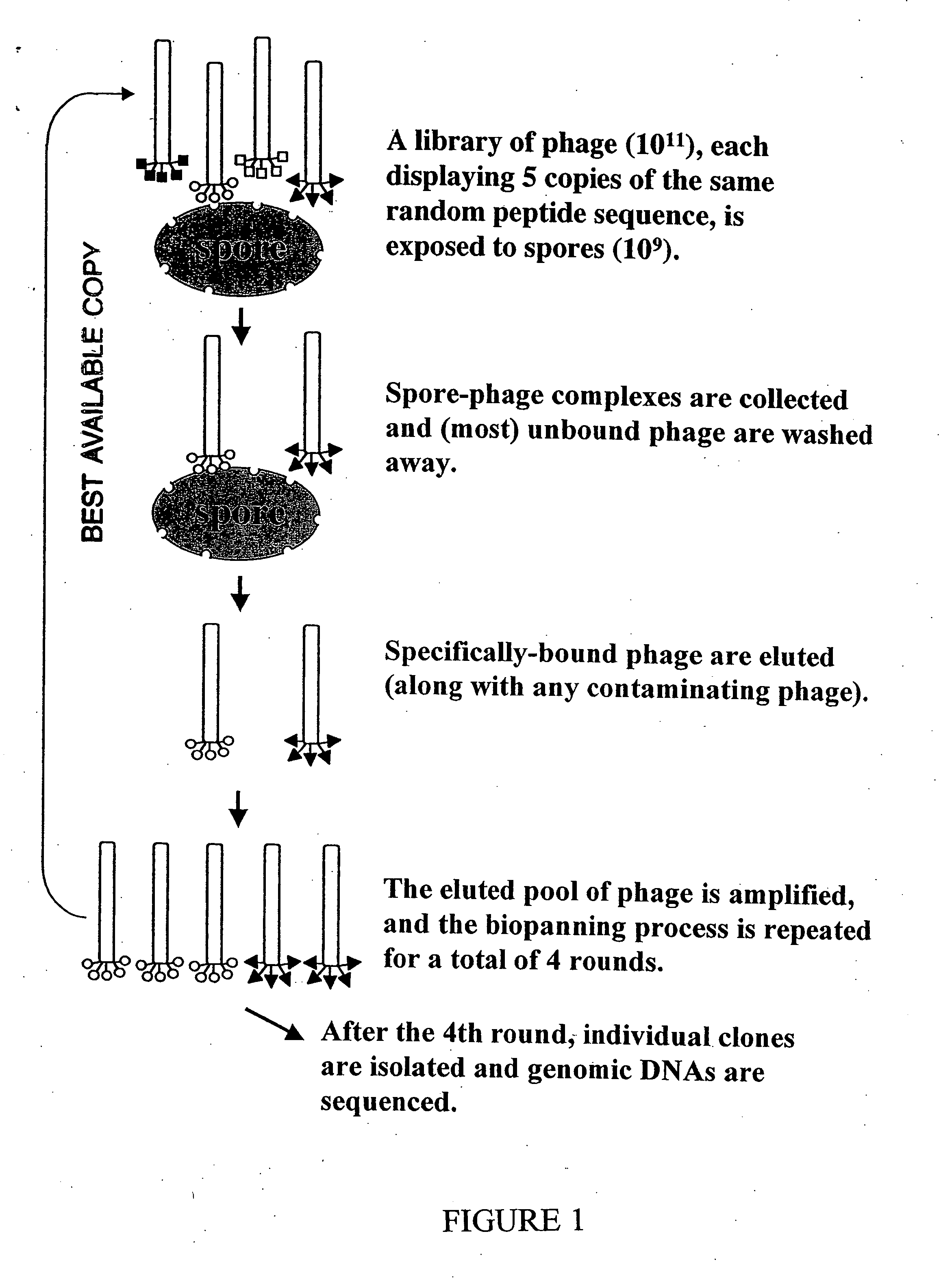
General Method of Biopanning of Phage Display Peptide Libraries with Bacillus Spores
[0036] Peptides of interest were identified using a phage display ligand screening system. A phage display peptide library kit (New England Bio Labs) was used according to instructions of the manufacturer in the identification process. These libraries display random 7- and 12-mer peptides, respectively, on the surface of the filamentous coliphage M13. These peptides are fused to the surface-exposed amino terminus of the minor capsid protein pIII, which is present in five copies at one end of the phage particle. Thus, each phage displays five copies of one particular peptide. The phage display peptide libraries, each contain approximately 2×109 independent phage clones. Each clone produces a protein pIII-peptide fusion that was created by the insertion of a random 21- or 36-base DNA fragment into a cloning site at the start of gene III of the phage. Consequently, the sequence of any peptide that bind...
example 2
Identification of B. subtilis-Specific Peptides
(a) Determination of the 7-mer Peptides
[0038] The DNA of over thirty independent phage isolates of B. subtilis were sequenced and thirteen unique sequences were identified. (See Table 1.) All encoded peptides contained amino terminal sequence Asn-His-Phe-Leu (SEQ ID No. 118). Although the sequences at positions five through seven are not identical, there is a clear preference for certain amino acids.
TABLE 1Nucleotide and Amino Acid Sequences fromB. subtilis Spore-Binding PhageIsolate 1AAT CAT TTT TTG ATT AAG CCG (SEQ ID No.2) 2AAT CAT TTT TTG AGG TCT CCG (SEQ ID No.3) 3AAT CAT TTT CTG CCT CGT TGG (SEQ ID No.4) 4AAT CAT TTT CTT CCT AAG GTG (SEQ ID No.5) 5AAT CAT TTT CTG TTG CCG CCG (SEQ ID No.6) 6AAT CAT TTT TTG CCT CCT CGG (SEQ ID No.7) 7AAT CAT TTT CTG CCT ACT GGG (SEQ ID No.8) 8AAT CAT TTT CTG ATG CCG AAG (SEQ ID No.9) 9AAT CAT TTT CTT AAG GGG ACG (SEQ ID No.10)10AAT CAT TTT TTG CCG CAG AAT (SEQ ID No.11)11AAT CAT TTT CTT CTT TGG...
example 3
Identification of B. anthracis-Specific Peptides
(a) Biopanning of B. anthracis-Specific Peptides with Avirulent ΔAmes Strain of B. anthracis
[0051] Biopanning with the heptamer phage display library was used to identify tight-binding peptides on the surface of B. anthracis spores. The spores were prepared from the avirulent delta-Ames strain of the organism (lacking the toxin-encoding plasmid pOX1) and were sterilized by gamma-irradiation by Diagnostics Systems Division of the U.S. Army Medical Research Institute of Infectious Disease, Fort Detrick, Md.
[0052] Four rounds of biopanning were performed in the manner described above. The genomic DNA of amplified elutes from each round were sequenced directly and genomic DNA of 27 single plaques from the fourth round amplified elute were also sequenced. In the fourth round of biopanning, the defining decrease in titer of the supernatant and the increase in the titer of the elutate indicated the selection of tight-binding phages. The D...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| wave lengths | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com