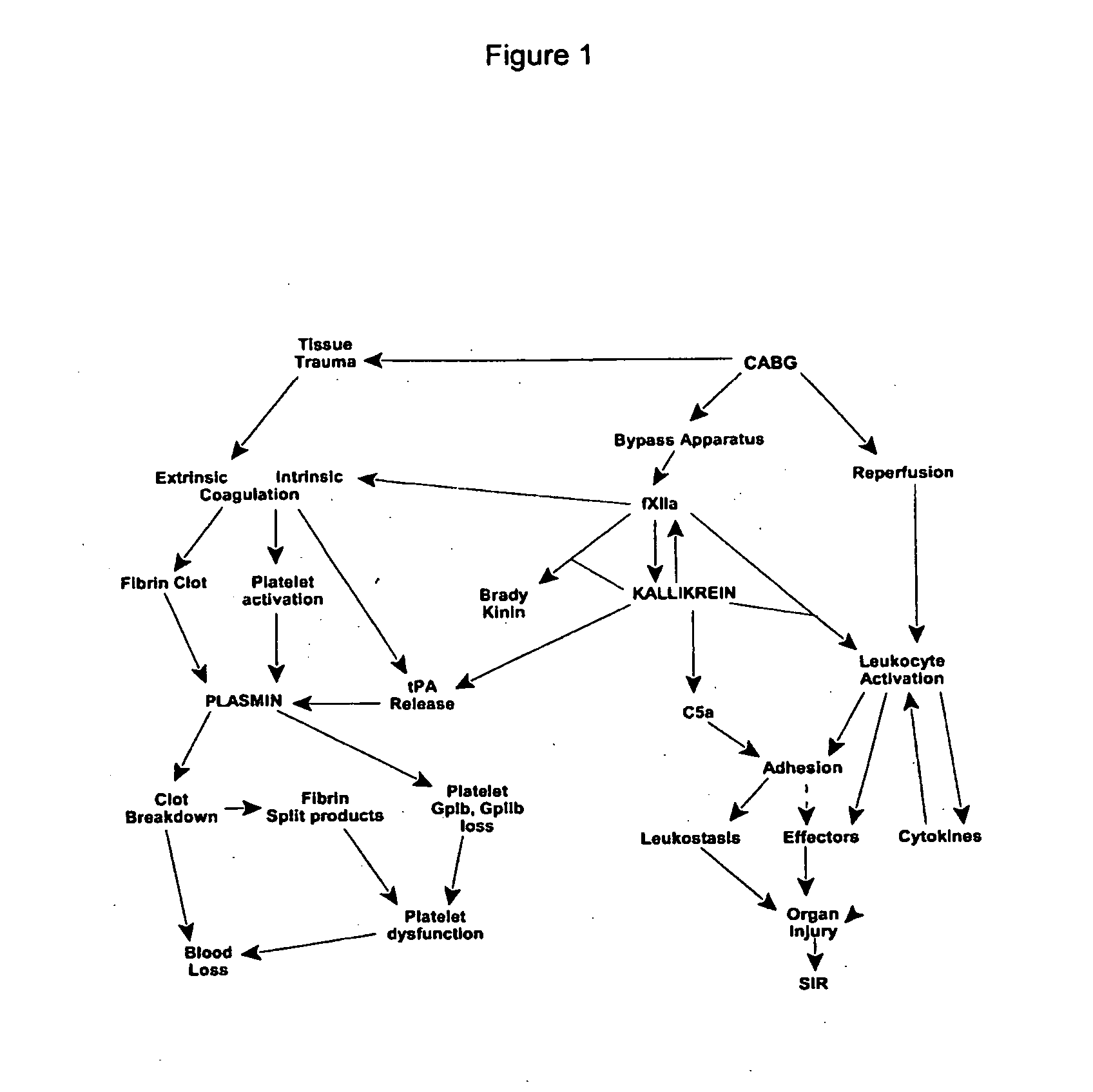
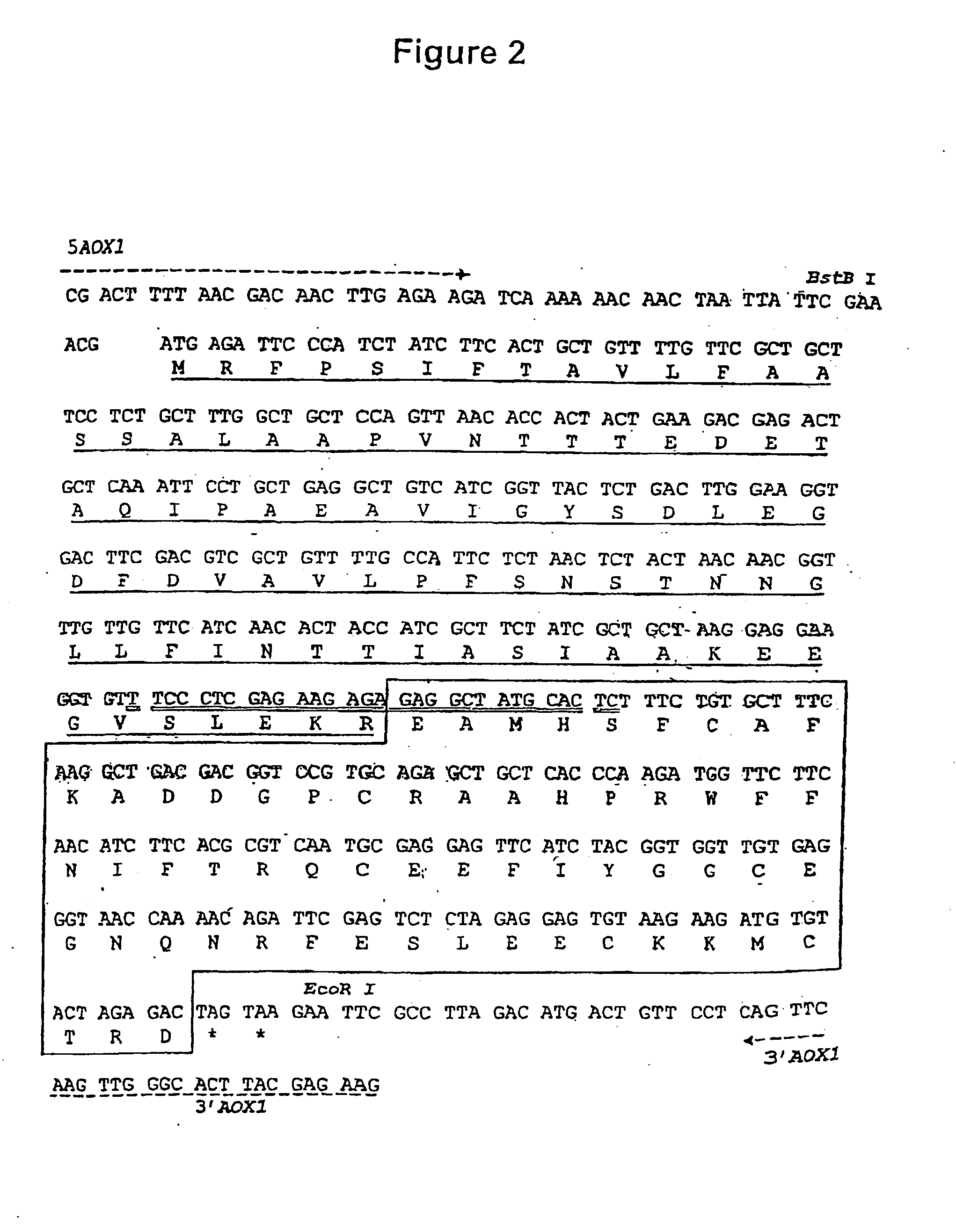
Prevention and reduction of blood loss
a technology of blood loss and prevention, applied in the direction of antinoxious agents, peptide sources, peptide/protein ingredients, etc., can solve the problems of contact activation, perioperative blood loss, etc., to prevent or reduce the onset of systemic inflammatory response, prevent or reduce the effect of ischemia
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
A Representative KI Polypeptide
[0064] A non-naturally occurring, KI polypeptide useful in the compositions and methods of the invention was identified as a kallikrein binding polypeptide displayed on a recombinant phage from a phage display library. PEP-1 has the following amino acid sequence: Glu Ala Met His Ser Phe Cys Ala Phe Lys Ala Asp Asp Gly Pro Cys Arg Ala Ala His Pro Arg Trp Phe Phe Asn Ile Phe Thr Arg Gln Cys Glu Glu Phe Ile Tyr Gly Gly Cys Glu Gly Asn Gln Asn Arg Phe Glu Ser Leu Glu Glu Cys Lys Lys Met Cys Thr Arg Asp (SEQ ID NO:2). The molecular weight of PEP-1 is 7,054 Daltons.
[0065] The nucleotide sequence (SEQ ID NO:3) encoding the PEP-1 amino acid sequence (SEQ ID NO:2), was derived from a peptide that was isolated and sequenced by standard methods determined from the recombinant phage DNA. PEP-1 was produced in amounts useful for further characterization as a recombinant protein in His4.sup.-phenotype host cells of yeast strain Pichia pastoris.
example 2
Construction of a Recombinant Plasmid to Express KI Polypeptides
[0066] The initial plasmid, pHIL-D2, is ampicillin resistant and contains a wild-type allele of His4 from P. pastoris. The final DNA sequence comprising the coding sequence for the mat.alpha. Prepro-PEP-1 fusion protein in the recombinant expression plasmid pPIC-K503 is shown in FIG. 2. The DNA sequence of pHIL-D2 was modified to produce pPIC-K503, as follows:
[0067] 1. The BstBI site in the 3′ AOX1 region of pHIL-D2, located downstream of the His4 gene, was removed by partial restriction digestion, fill-in, and ligation, altering the sequence from TTCGAA (SEQ ID NO:23) to TTCGCGAA (SEQ ID NO:24). This modification was made to facilitate and direct the cloning of the expression cassette into the plasmid.
[0068] 2. The AatII site bearing the bla gene located downstream of His4 was removed by restriction digestion, fill-in, and ligation modifying the sequence from GACGTC (SEQ ID NO:25) to GACGTACGTC (SEQ ID NO:26). This ...
example 3
Manufacture of PEP-1 from Recombinant Yeast Cell Line
[0070] Spheroplasts of P. pastoris GS115 having the His4.sup.-phenotype were transformed with the expression plasmid pPIC-K503 (above) following linearization of the plasmid at the SacI site and homologous recombination of the plasmid DNA into the host 5′ AOX1 locus. The phenotype of the production strain is His4.sup.+. The entire plasmid was inserted into the 5′ AOX1 genomic sequence of the yeast.
[0071] Isolates from the transformation were screened for growth in the absence of exogenous histidine with methanol as the sole carbon source. Greater than 95% of the transformants retained the wild-type ability to grow with methanol as the sole carbon source, thereby demonstrating that the plasmid had been inserted into the host genome by homologous recombination rather than transplacement. These transformants did not require exogenous histidine for growth, thereby demonstrating that the plasmid had integrated into the host genome. S...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com