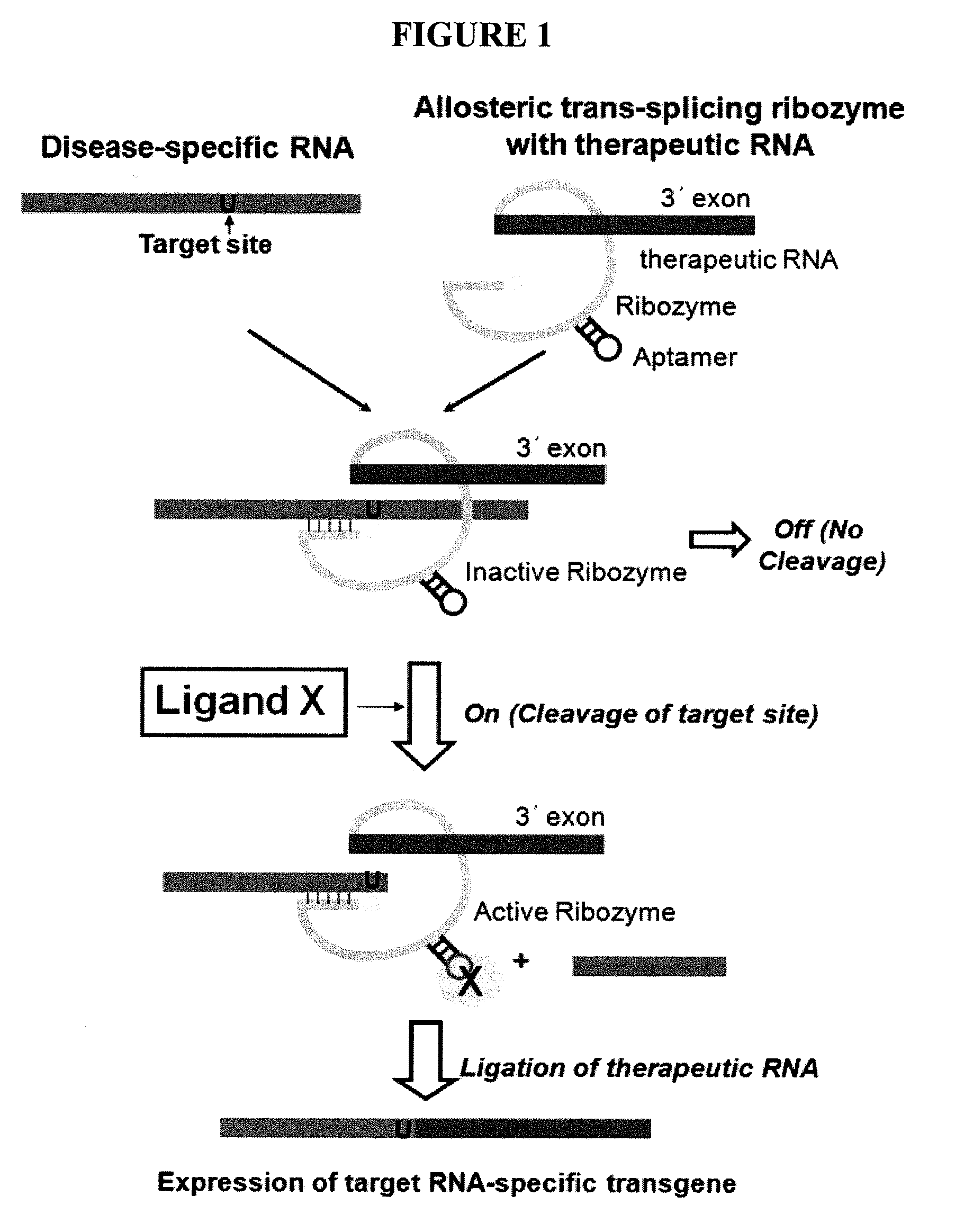
Allosteric trans-splicing group i ribozyme whose activity of target-specific RNA replacement is controlled by theophylline
a technology of allosteric transsplicing and ribozyme, which is applied in the field of allosteric transsplicing group i ribozyme whose target, can solve the problems of not maximizing the desired effect of treatment diseases, many problems to be solved in the existing gene therapy technology, and the inability to maximize the therapeutic effect of diseases,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
reference example 1
Preparation of Substrate (hTERT) RNA
[0070]To prepare target RNA, a pCl-neo vector (exon 1-2) containing a −1st to +218th DNA sequence of the hTERT was PCR-amplified with a primer (5′-GGGGAATTCTAATACGACTCACTATAGGGCAGGCAGCGCTGCGTCCT-3′) set forth in SEQ ID NO: 9 and a primer (5′-CGGGATCCCTGGCGGAAGGAGGGGGCGGCGGG-3′) set forth in SEQ ID NO: 10, thus to prepare a DNA fragment encoding hTERT RNA. The DNA fragment thus prepared was transcribed in vitro into RNA. A DNA template (3 μg), a 10× transcription buffer, 10 mM DTT (Sigma), 0.5 mM ATP, GTP, CTP and UTP (Roche), an 80U RNase inhibitor (Kosco), a 200U T7 RNA polymerase (Ambion) were added, and DEPC-H2O was added to a final volume of 100 μl, and then mixed. Then, the resulting mixture was reacted at 37° C. for 3 hours, and further treated with 5U DNase I (Promega) at 37° C. for 30 minutes to completely remove the DNA template. RNA was purified through the phenol extraction (pH 7.0) and ethanol precipitation, and separated on 6% denatur...
reference example 2
Cloning of Theophylline-Dependent hTERT Targeting Trans-Splicing (T / S) Aptazyme
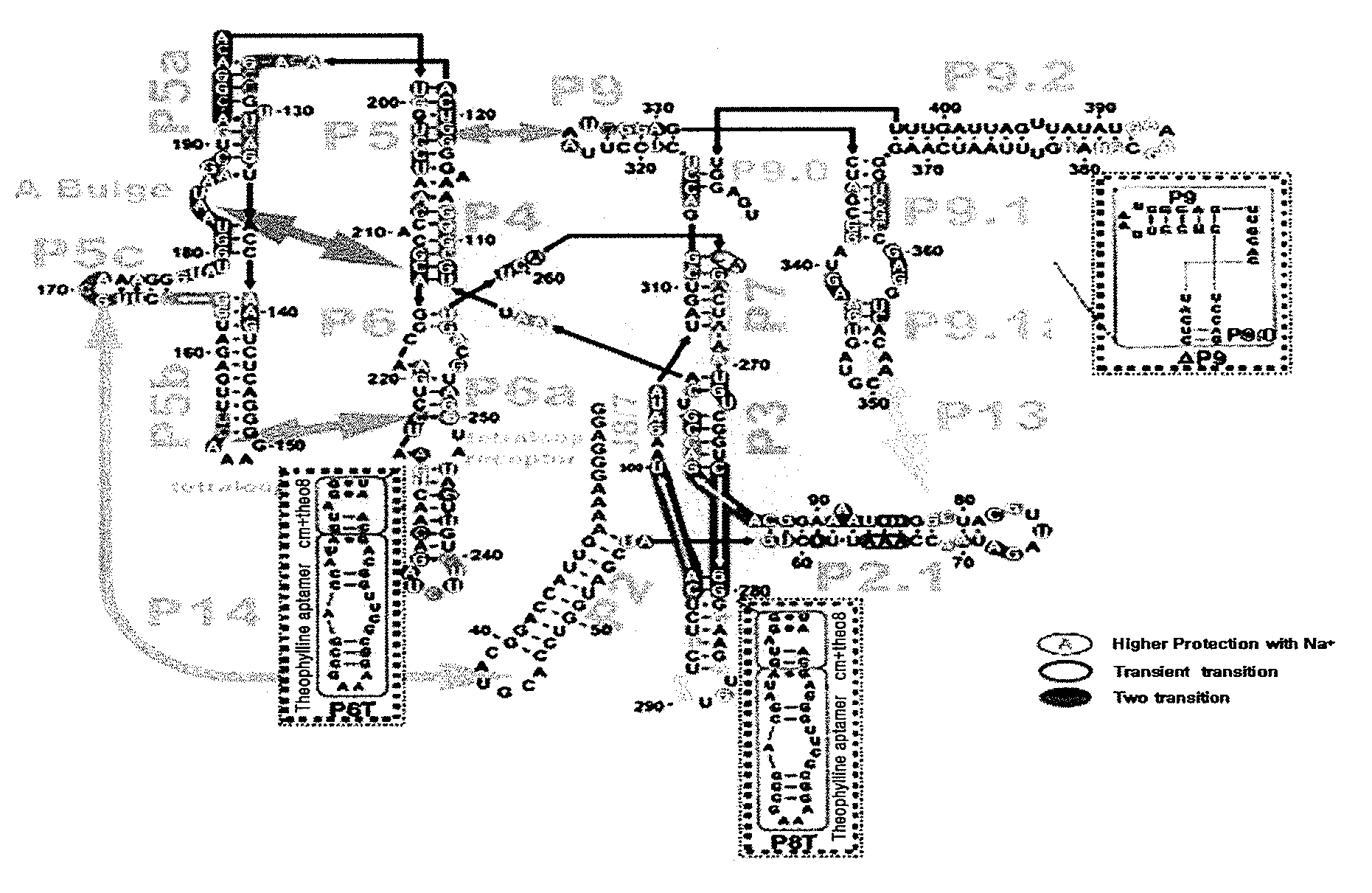
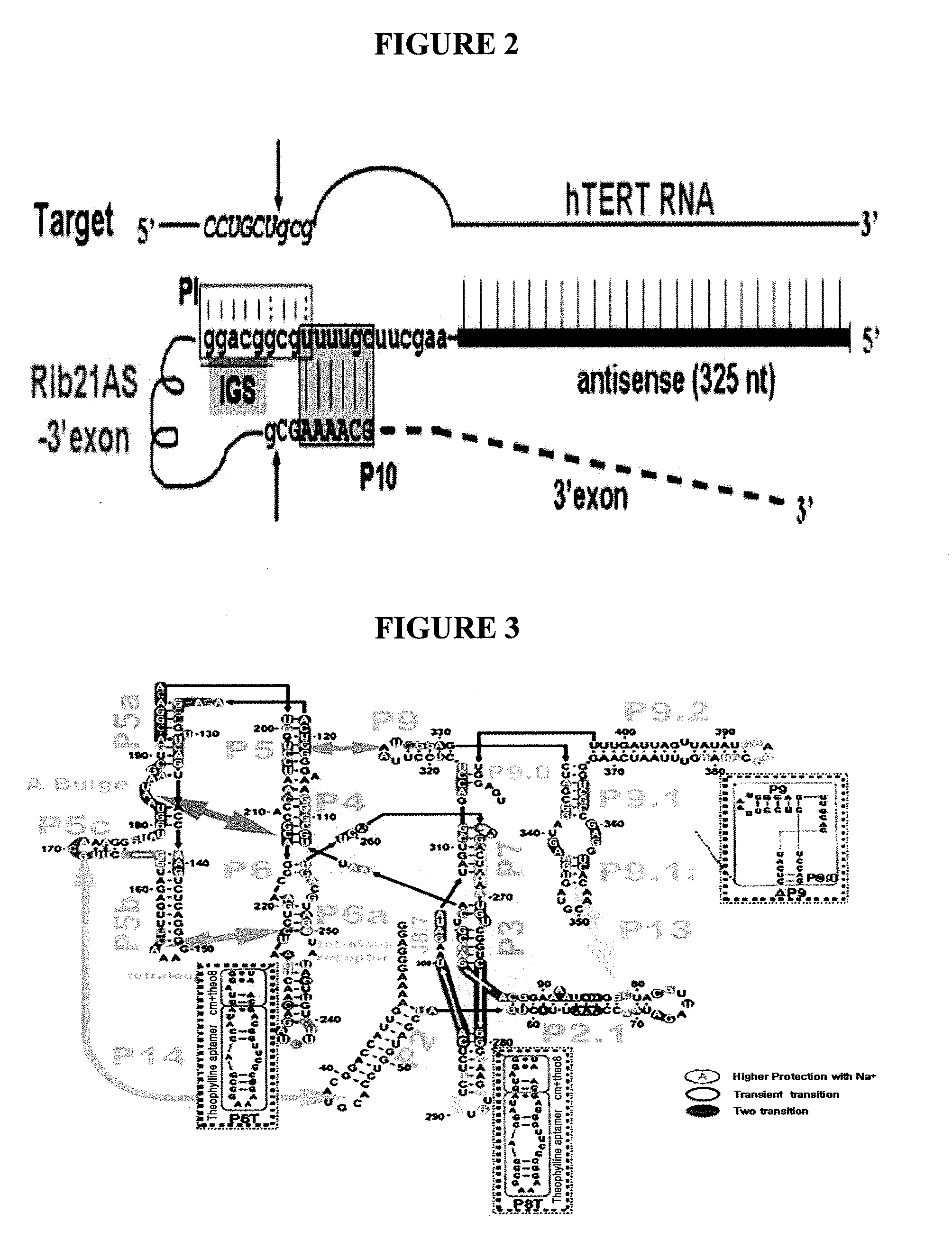
[0071]As a basic trans-splicing ribozyme backbone used to develop an allosteric ribozyme, group I intron ribozyme, which specifically recognizes a +21 nt site of hTERT and has P1, P10 and extended IGS to which 300 nt anti-sense sequence against target RNA is annealed, was used (Kwon, B. S., Jung, H. S., Song, M. S., Cho, K. S., Kim, S. C., Kimm, K., Jeong, J. S., Kim, I. H., and Lee, S. W. 2005, Specific regression of human cancer cells by ribozyme-mediated targeted replacement of tumor-specific transcript. Mol. Ther. 12: 824-834; Hong, S. H., Jeong, J. S., Lee, Y. J., Jung, H. I., Cho, K. S., Kim, C. M., Kwon, B. S., Sullenger, B. A., Lee, S. W.*, and Kim, I. H.* 2008, In vivo reprogramming of hTERT by trans-splicing ribozyme to target tumor cells. Mol. Ther. 16: 74-80).
[0072]A theophylline aptamer was cloned into either or both of P6 and P8 domains of the hTERT targeting ribozyme by means of a communica...
reference example 3
Preparation of Theophylline-Dependent hTERT Targeting T / S Aptazyme RNA
[0074]The DNA sequence of the theophylline-dependent hTERT targeting T / S aptazyme prepared in Reference example 2 was PCR-amplified with a primer set forth in SEQ ID NO: 16 (5′-GGGGAATTCTAATACGACTCACTATAGGCAGGAAAAGTTATCAGGCA-3′) including a T7 polymerase promoter and a primer set forth in SEQ ID NO: 17(5′-CCCAAGCTTGCGCAACTGCAACTCCGATAA-3′) that is annealed with the midway site of the 3′ exon of the ribozyme. In this case, an increased amount of a DNA template (3 μg) and NTP (1.5 mM) was used to prevent a self-splicing reaction as much as possible. Then, a 1× splicing buffer (40 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 10 mM DTT and 4 mM spermidine), 0.5 mM ATP, GTP, CTP, UTP (Roche), an 80U RNase inhibitor (Kosco), a 200U T7 RNA polymerase(Ambion) were added, and DEPC-H2O was added to a final volume of 100 μl and then mixed. Then, the resulting mixture was transcribed at 37° C. for 3 hours, and further treated with 5U DNa...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molar density | aaaaa | aaaaa |
| Gene expression profile | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com