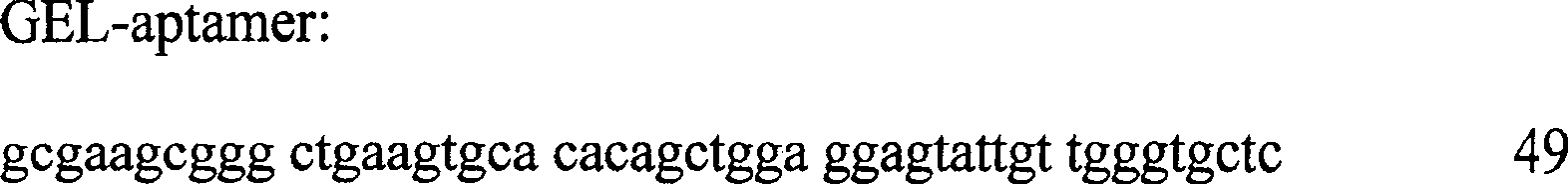Method for detecting protein or protein complex by identifying a plurality of epitopes synchronously
A protein and epitope technology, applied in the field of proteomics research, can solve the problems of cumbersome, high GC content of oligonucleotide aptamers, extension of auxiliary fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
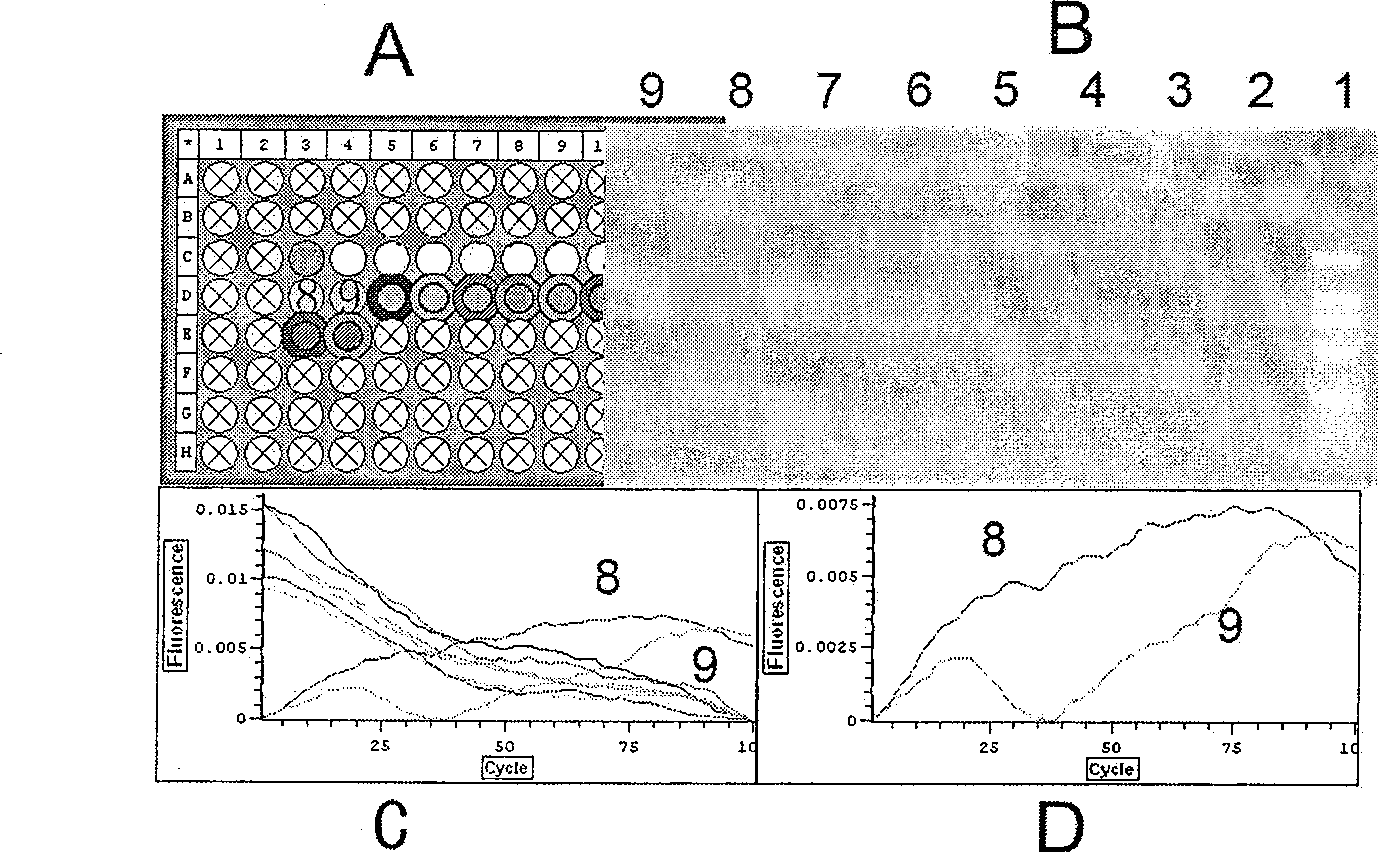
[0049] Four oligonucleotide aptamers detect CatD protein
[0050] 1. Add 1.0uL 100pM four kinds of oligonucleotide aptamer mixture into the PCR tube
[0051] 2. Dilute the CatD protein with 1% BSA as the diluent, then add the CatD protein with a concentration of 1.0aM, 100.0aM and 10.0fM and 1% BSA as a control in different tubes, and place it at room temperature for 1 hour
[0052] 3. Add 0.1uL 25uM four kinds of linker mixture to the buffer containing 1.0U T4 DNA Ligase, add this mixture to the reaction tube and place it at room temperature for 5 hours for ligation reaction
[0053] 4. Add 1.0uL 10X phi29 buffer, 0.5uL 10mM dNTP, 0.2uL 100X BSA, 0.3uL phi DNA polymerase, 7.5uL ddH to each tube 2 O, 0.5 uL Eva Green, 0.1 uL primer1 and 0.1 uL primer2. Amplify at 30°C for 8-12 hours
[0054] 5. Start to use OPTICON2 continuous fluorescence detector (MJ Research) to record data for four hours at the 8th hour, read the value every 2.5 minutes, and last for about four hours, mai...
Embodiment 2
[0057] Four oligonucleotide aptamers detect CatD protein
[0058] 1. Add 1.0uL 100pM four kinds of oligonucleotide aptamer mixture into the PCR tube
[0059] 2. Dilute the CatD protein with 1% BSA as the diluent, and then add CatD protein with a concentration of 1.0aM, 100.0aM and 10.0fM and 1% BSA as a control in different tubes; CatD protein with three epitopes was used as a control, that is, 10.0 nM and 1.0 nM proteins were added to the tube, and left at room temperature for 1 hour
[0060] 3. Add 0.1uL of 25uM four kinds of linker mixture to the buffer containing 1.0U of T4 DNA Ligase, add this mixture to the reaction tube and place it at room temperature for 7 hours to carry out the ligation reaction
[0061] 4. Add 1.0uL 10X phi29 buffer, 0.5uL 10mM dNTP, 0.2uL 100X BSA, 0.3uL phi DNA polymerase, 8.0uL ddH to each tube 2 O, 0.1uL primer1 and 0.1uL primer2 were amplified at 30°C for 36 hours
[0062] 5. Use 2% agarose electrophoresis for detection
[0063] 6. The resu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com