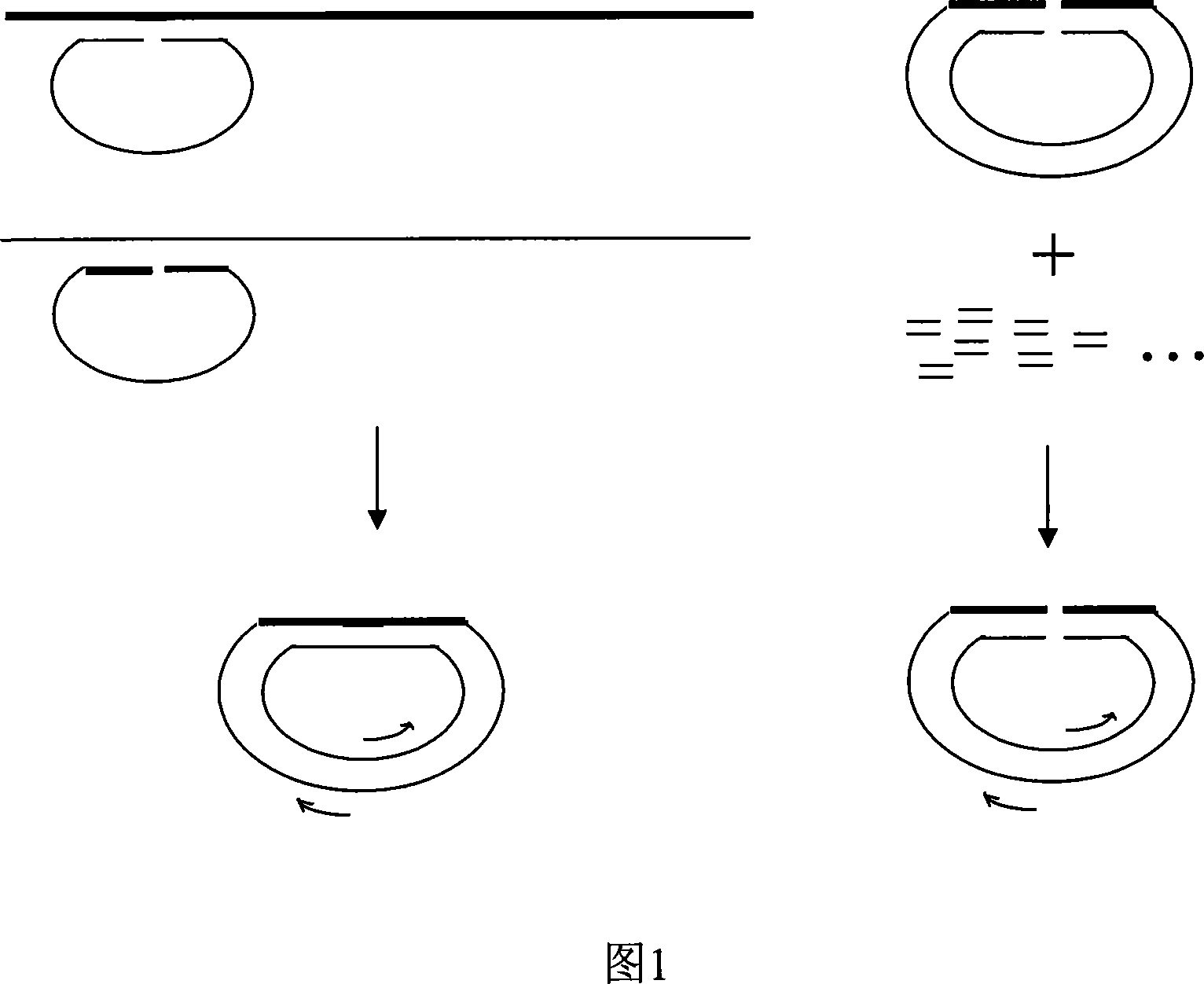
Systemic displacement polymerase lymerase chain reaction
A polymerase chain reaction technology, which is applied in the field of system replacement polymerase chain reaction, can solve the problems of high false positives and easy PCR contamination.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0086] Embodiment. Hepatitis B HBsAg gene detection:
[0087] (1) Purification of DNA from 11 clinical specimens (according to Qiagen instructions):
[0088] 1): Add 100-200ul sample to a 1.5ml EP tube, add an equal amount of phenol / chloroform / isoamyl alcohol (25:24:1) for extraction, vortex, and centrifuge at the highest speed for 5 minutes in a desktop centrifuge.
[0089] 2): Transfer 100-200ul of the supernatant to a new EP tube, add an equal amount of chloroform, shake, and centrifuge.
[0090] 3): Add 4 times Qiagen binding buffer to the supernatant and transfer to the purification column, wash the column twice with the washing buffer, add 50ul dH 2 Purified samples were collected by O elution.
[0091] (2) Indicates the preparation of template tailed DNA PCR:
[0092] 1) Select the gene sequence of hepatitis B virus HBs Ag aa124-138
[0093] 5’-gc acg att cct gct caa gga acc tct atg ttt ccc tct tg-3’
[0094] 3'-cg tgc taa ggt cga gtt cct tg-5' as the reserved sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com