Polypeptide having a phytase activity and nucleotide sequence coding therefor
A technology of phytase activity and phytase, applied in the field of polypeptides with phytase activity and nucleotide sequences encoding polypeptides, can solve problems such as limited characteristics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Embodiment 1: the mensuration of phytase activity
[0086] Phytase activity was detected with a detection mixture consisting of 0.5% phytic acid (about 5 mM), 200 mM sodium citrate, pH 5.0. After co-incubating at 37°C for 15 minutes, the same volume of 15% trichloroacetic acid was added to stop the reaction. Mix 100 microliters of the detection mixture with 900 microliters of water and 1 milliliter of 0.6M sulfuric acid, 2% ascorbic acid and 0.5% ammonium molybdate, react at 50°C for 20 minutes, and quantitatively measure the released phosphorus ions at 820nm . A standard solution of potassium phosphate was used as a reference.
Embodiment 2
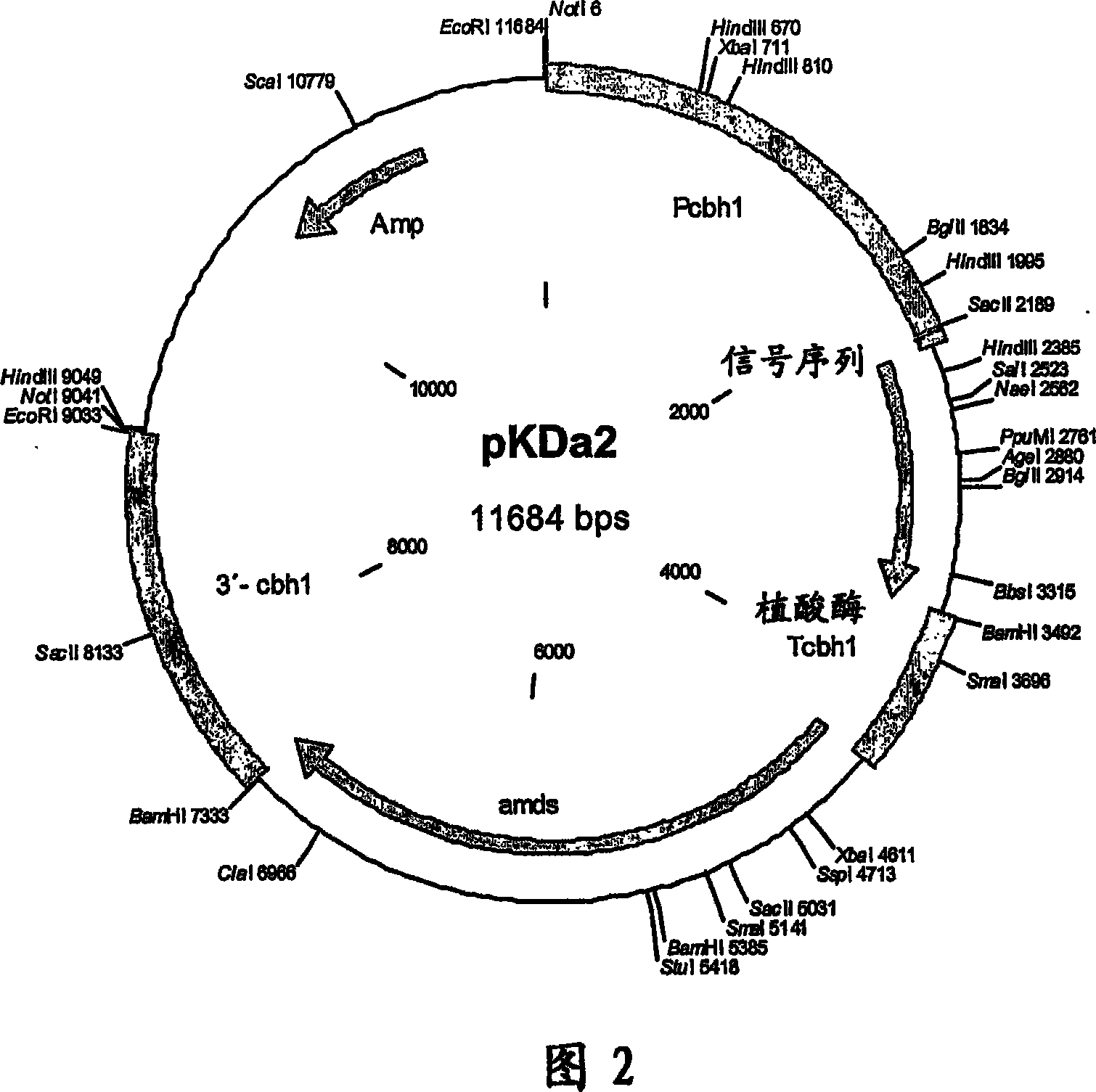
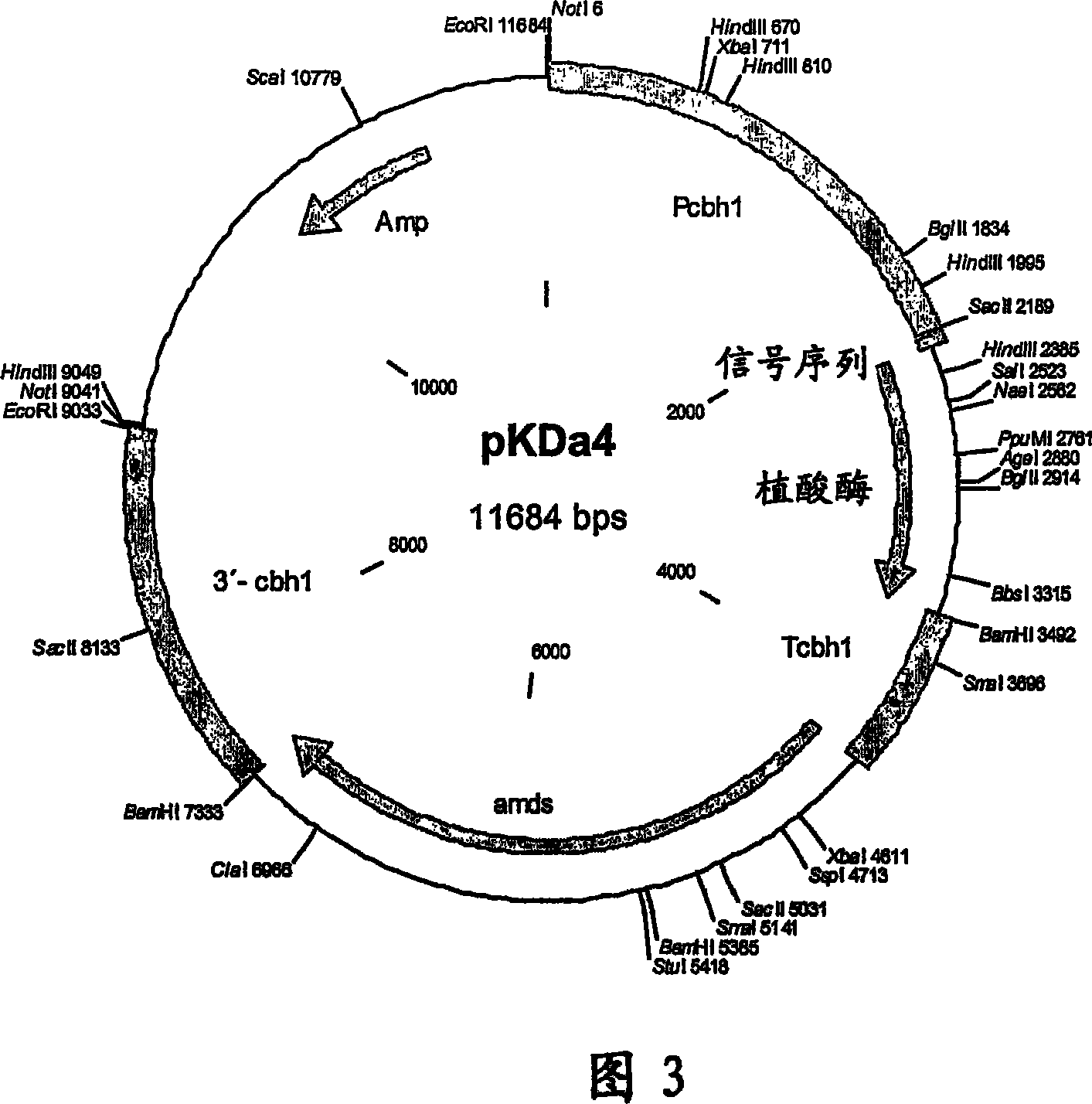
[0087] Example 2: Construction of pKDa2 and pKDa4 plasmids
[0088] The sequence encoding the phytase from E. coli (Dassa et al. 1990, J. Bacteriol. 172: 5497-5500, accession number: M58704) utilizes codons from Trichoderma reesei (http: / / www.kazusa.or.jp / codon) generation and synthesis. All synthetic fragments were sequenced and the fragments with and without mutations were joined together to generate new phytase variants. In one of the resulting variants, the amino acid Val of the phytase gene 200 (GTG) becomes Tyr 200 (TAC). This variant is called Da2. The original polynucleotide ("Stammpolynucleotid") without mutations was called Da4. Mature E. coli phytase gene clones were amplified by PCR. The DNA sequence containing the CAG (Gln) codon at position 1 contains an open reading frame of 1,230 bp, which encodes a phytase of 410 amino acids (SEQ ID NO: 1 / 2).
[0089] The signal peptide of the A. niger phytase (SEQ ID NO: 3 / 4) was used to enable secretion of the E. col...
Embodiment 3
[0101] Example 3: Transformation of Trichoderma reesei with pKDa2 and pKDa4 to obtain single copy transformants
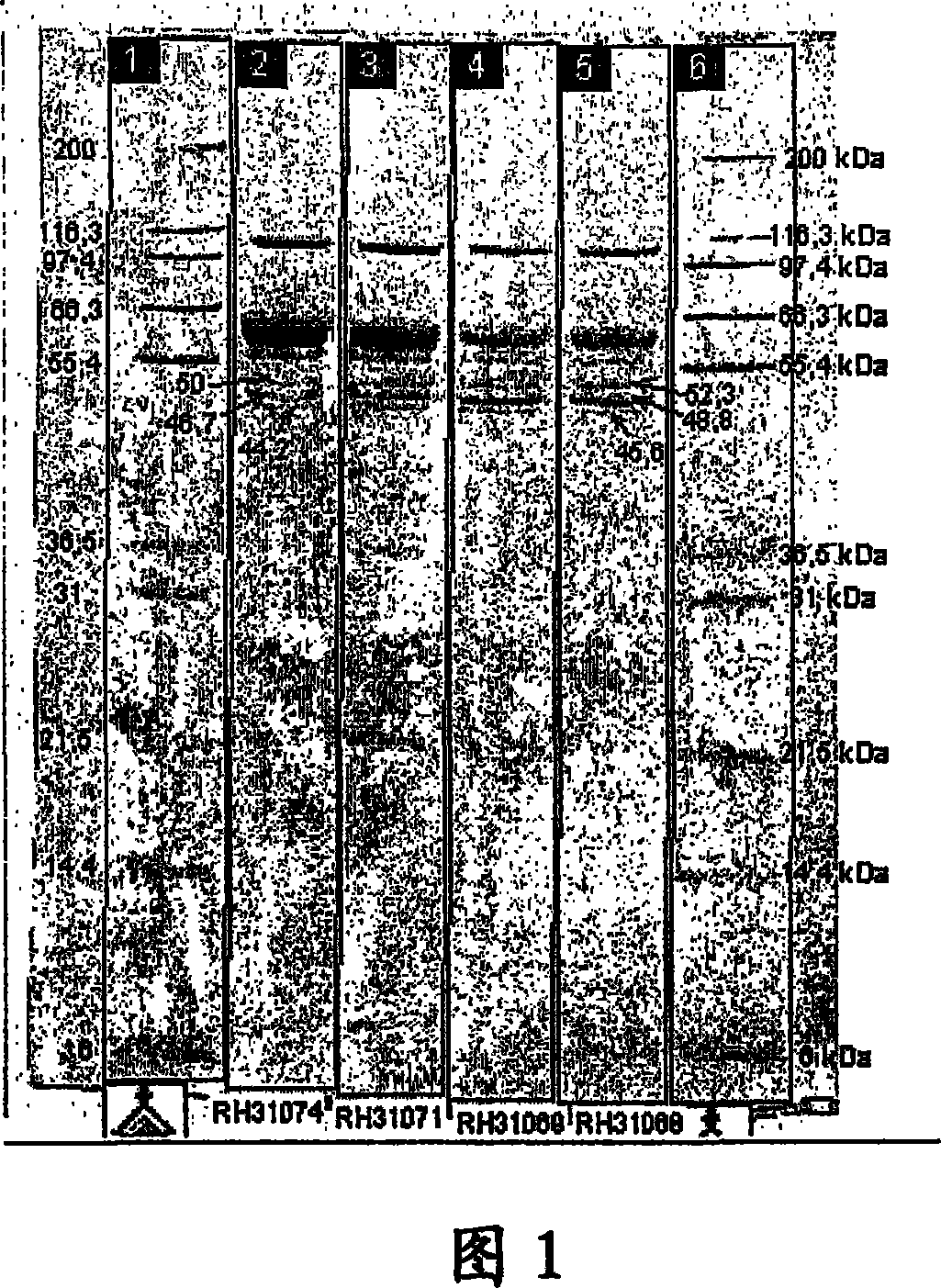
[0102] Trichoderma reesei RH 3780d was transformed with linearized expression cassettes isolated from plasmids pKDa2 and pKDa4, respectively. The technique for transforming and maintaining Trichoderma reesei refers to the technique published by Penttil et al. (1987, Gene 61: 155-164). Transformants were screened and purified twice by single spore isolation. Those transformants with the highest secretion efficiency were selected from all transformants. These transformants containing DNA from plasmid pKDa2 were designated RH 31068 and RH 31069, and those containing DNA from plasmid pKDa4 were designated RH 31071-31075 and were used for further characterization.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com