Auxiliary screening method for indica rice and japonica rice
An indica rice and japonica rice technology, which is applied in the directions of biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., to achieve the effect of optimizing hybrid combinations, low cost, and quick selection methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
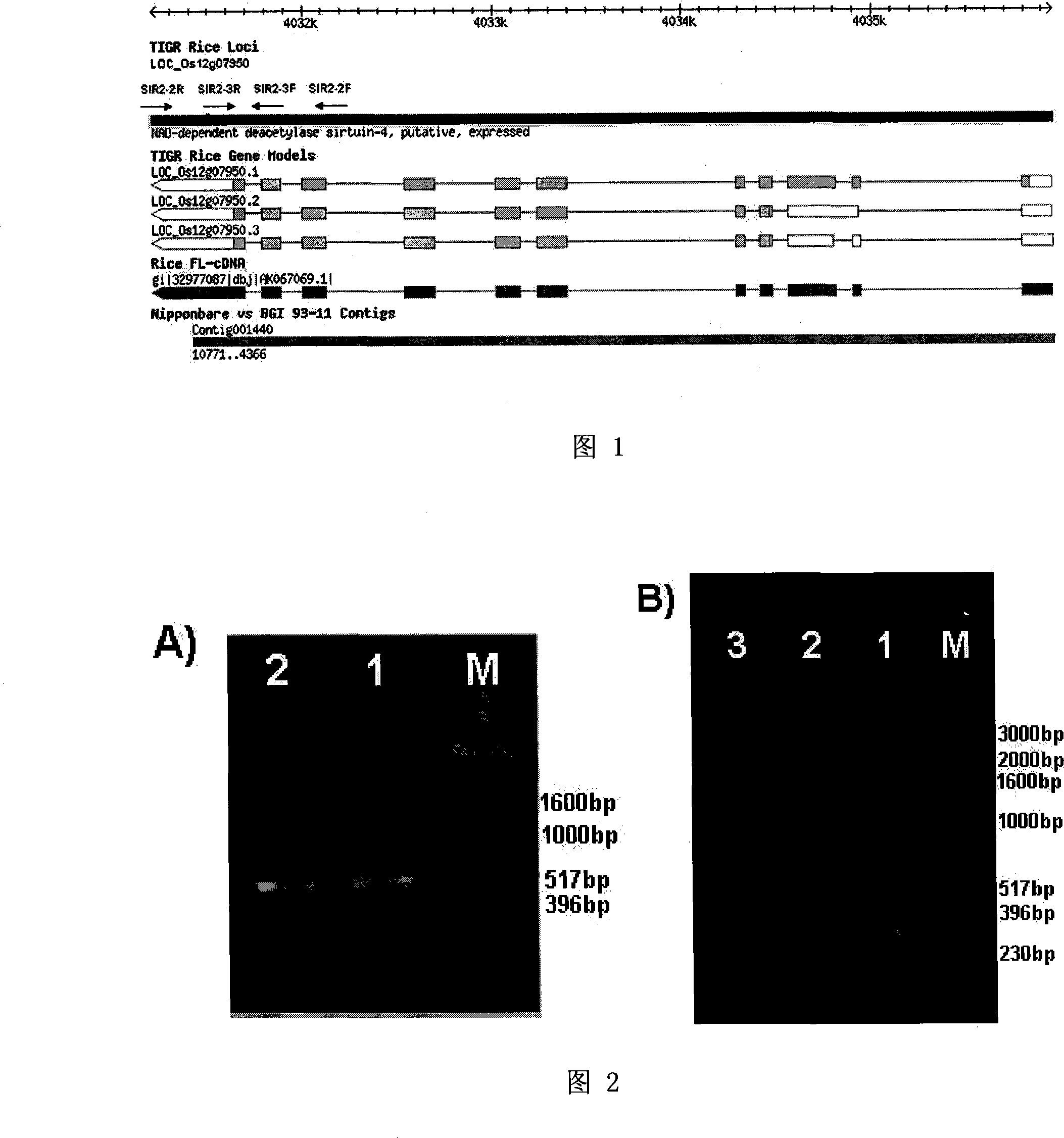
[0020] Example 1. Discovery and verification of different expression patterns of gene LOC_Os12g07950 in indica and japonica rice
[0021] Firstly, by mining and analyzing the chip data of indica and japonica rice at different developmental stages and different tissue parts, it was found that the gene LOC_Os12g07950 had two sets of probes in the GengChip data, and the expression patterns of these two sets of probes in indica and japonica rice were opposite. , the expression level of probe P1 is very low or even not expressed in indica rice, but the expression level in japonica rice is very high, while the expression level of another probe P2 is very high in indica rice, but very low in japonica rice . According to previous research results, the main reason for the difference in gene expression may be the difference in base sequence (InDel), or it may be regulated by other genes. Therefore, using the method of comparative genomics, the genome sequence of gene LOC_Os12g07950 in ...
Embodiment 2
[0022] Embodiment 2, be used for the design of the primer of auxiliary screening indica rice and japonica rice
[0023] In order to further verify whether there is a genome-level difference between 93-11 and Nipponbare, according to the Nipponbare sequence, primers SIR2-2 were designed in the region where there may be genomic differences between the two. One of the primer sequences is shown in sequence 1 in the sequence table, and the other A primer sequence is shown as sequence 2 in the sequence table, and the genomic DNA of Nipponbare and 93-11 were respectively used as templates for PCR amplification verification. The reaction system of PCR amplification is: rice genomic DNA template 20ng, Taq Plus DNA polymerase 0.5U, 2.0μl 10×PCR buffer (100mM Tris HCl pH9.0, 500mM KCl, 15mM Mg 2+, 1% Triton X-100), 100 μM dNTPs, 4 μM forward primer, 4 μM reverse primer, supplement the reaction system with DEPC water to 20 μl. The PCR reaction conditions were as follows: first at 94°C fo...
Embodiment 3
[0025] Example 3. Feasibility verification of using primer SIR2-3 to assist in screening rice indica and japonica subspecies
[0026] Rice varieties and local species (both from the National Germplasm Bank): 93-11, Xima Zhagu, Zinuo, Nepa (deep water rice), Guichao 2, Aus6538 (Bamalaia), IR24, Teqing, Nipponbare, Ai Dazhong, Muli (japonica rice with light shell), glutinous japonica rice, Yuefu, Qiuguang, Hongguang and Yunfeng 7.
[0027] First, 8 varieties and landraces of each subspecies of indica and japonica were randomly selected by using morphological indicators and related literature retrieval, as shown in Table 1:
[0028] Table 1. Identification results of indica and japonica subspecies of 16 rice varieties and landraces
[0029] species name
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com