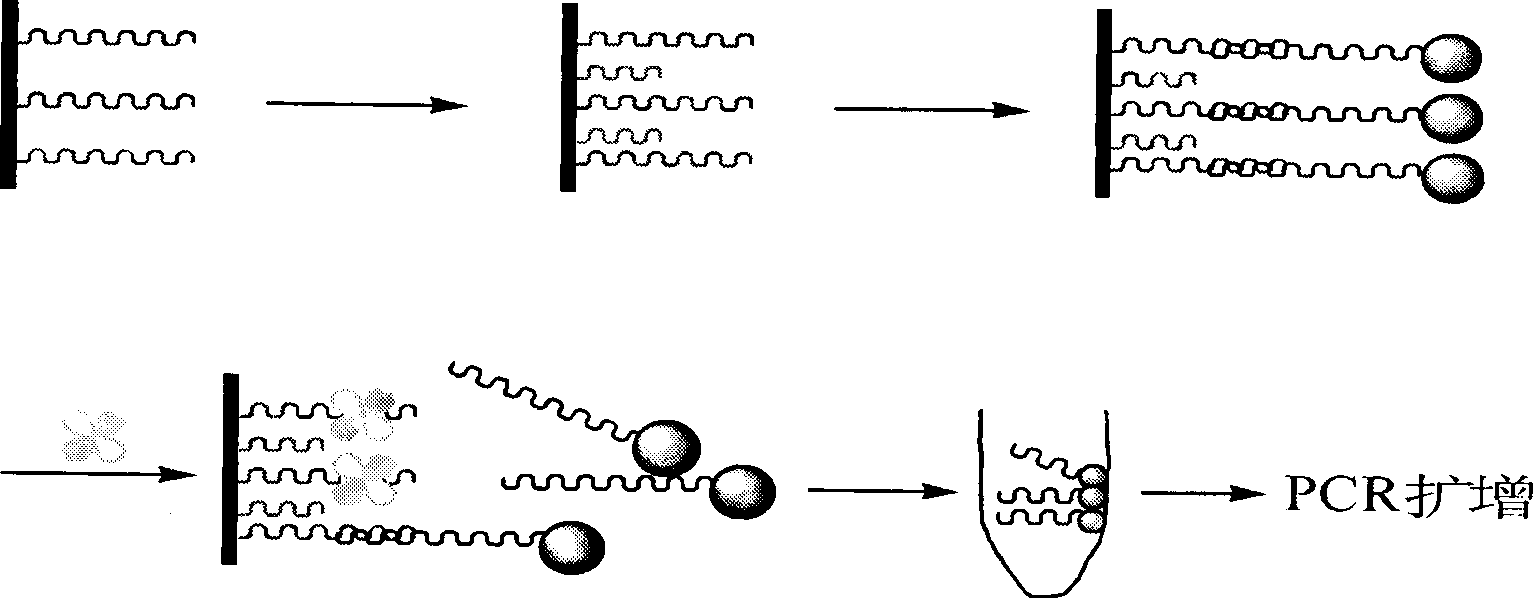
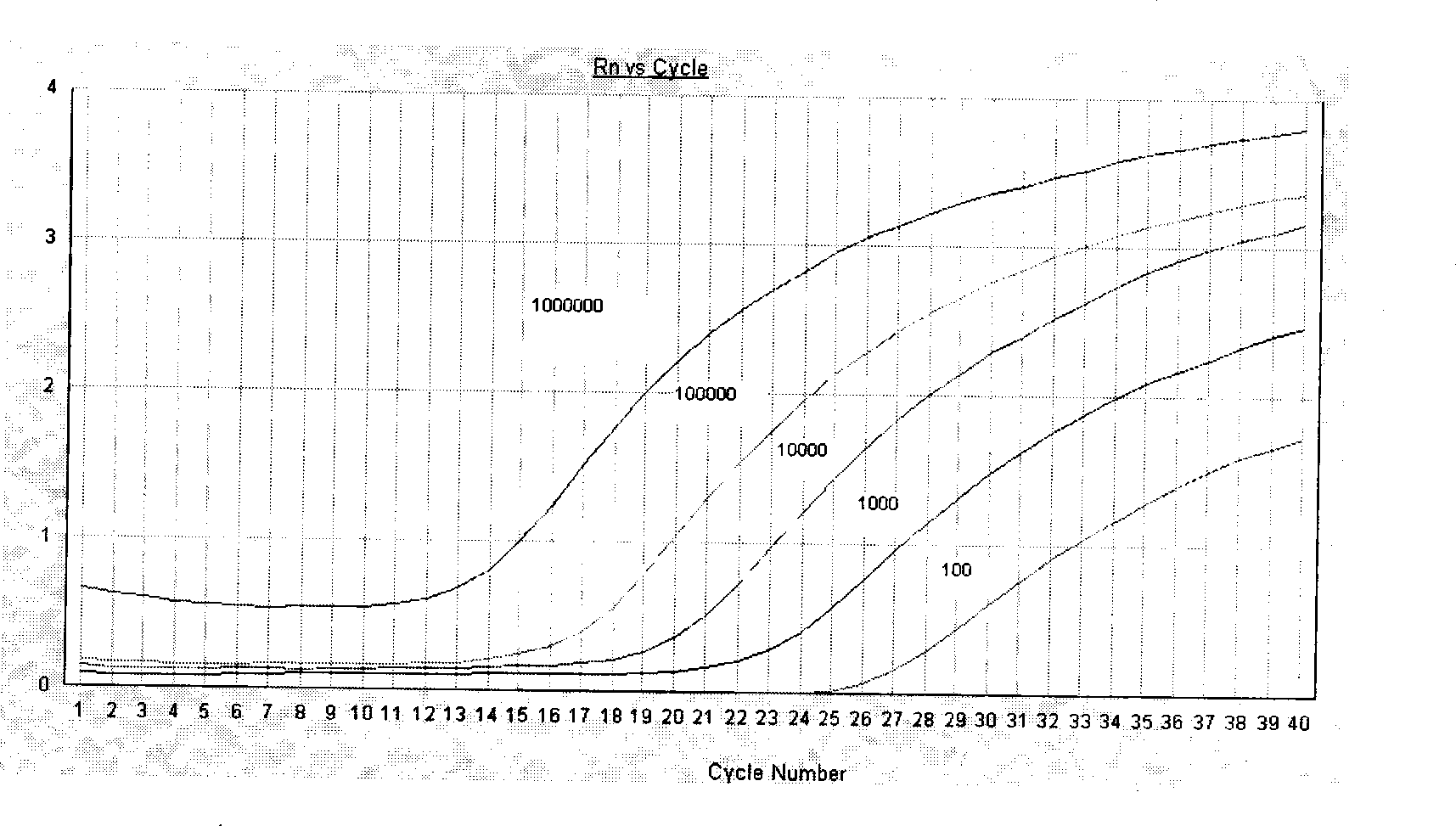
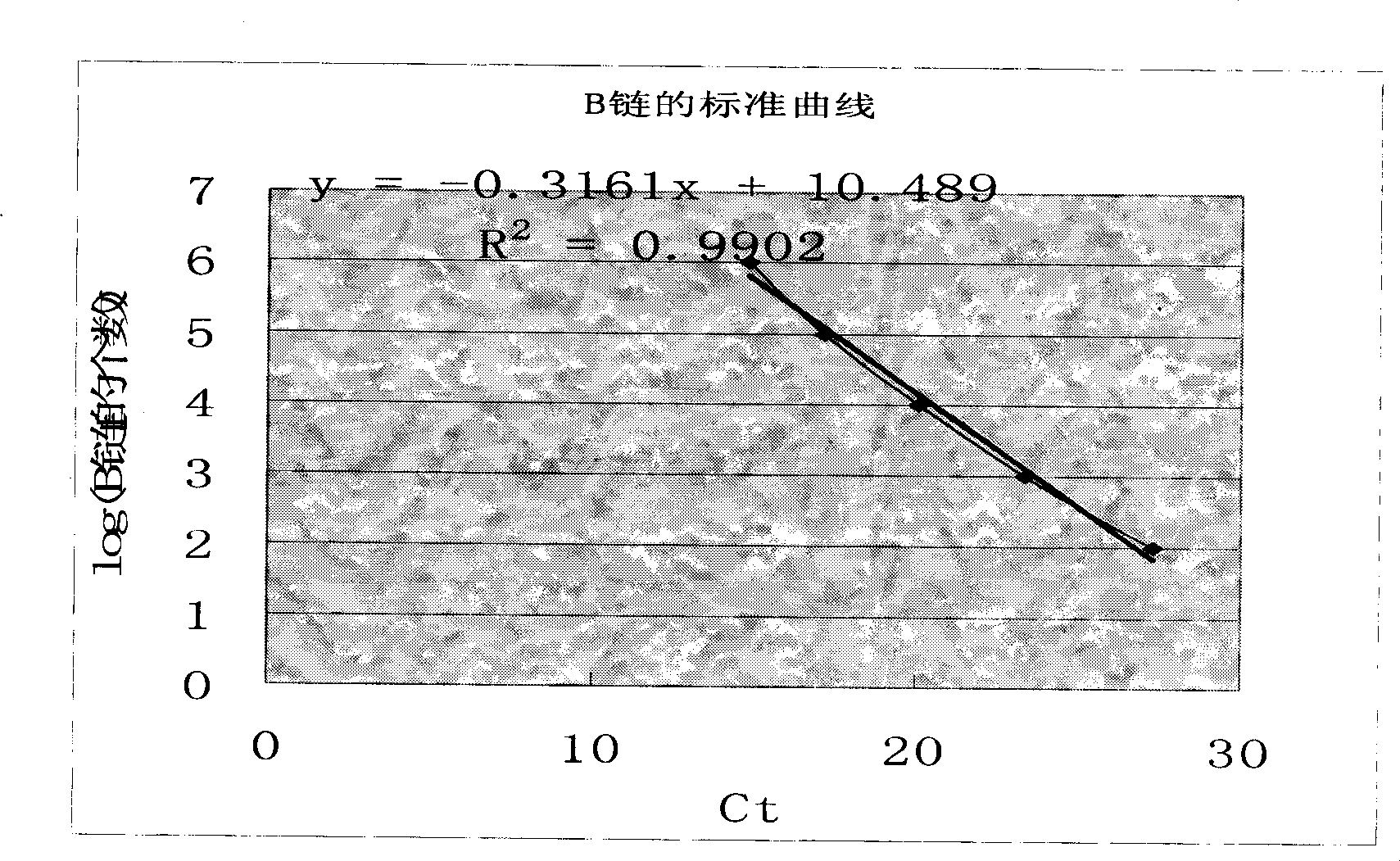
Method for detecting target numerator based on nucleic acid aptamer and PCR amplification
A nucleic acid aptamer and target detection technology, which is applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of limited application, achieve the effect of few influencing factors, simple operation and steps, and reliable results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Example 1: Use Aptamer based PCR to detect trace amounts of cocaine in serum or plasma (small molecule detection) (without magnetic bead enrichment and separation)
[0033] DNA sequences used in this experiment:
[0034] A chain sequence: (39 bases)
[0035] 3-HS-(CH2) 6 -CTG GGT GAA GTA ACT TCC TAA AAG GAA CAG AGG GGGAGT-5
[0036] B chain sequence: (58bases)
[0037] 5-GTGTGAAGCGGCATCTAAAGCGTCTCCCCCCTCAAGACTAGGTCCAGCCCAGGGTGTC-biotin-3
[0038] The part in italics is the hybridization site of the template
[0039] B chain primer:
[0040] S: 5-GTG TGA AGC GGC ATC TAA AGC-3 (21bases)
[0041] A: 5-GAC ACC TCT GGG CTG GAC C-3 (19bases)
[0042] Experimental steps:
[0043] 1. Treatment of gold electrodes
[0044] 1. First drop a drop of separation reagent on the gold electrode, after 15 minutes, wash it off with double distilled water.
[0045] Separation reagent: H 2 o 2 : Concentrated H 2 SO 4 =7:3 (volume ratio)
[0046] 2. Use Al 2 o 3The slurry pol...
example 2
[0089] Example 2: Using Aptamer based PCR to detect thrombin in serum (protein detection)
[0090] DNA sequences used in this experiment:
[0091] A chain sequence:
[0092] 5-HS-(CH2) 6 -ATATAGGTTGGTGTGGTTGG-3
[0093] B chain sequence:
[0094] 5-CAG TGA AGC GGC ATC TAA AGC ACG AGT CTG AGT CCA ACC ACA-3
[0095] Primer for B chain: A: 5-TGT GGT TGG ACT CAG AC TCG-3
[0096] S: 5-GTG TGA AGC GGC ATC TAA AGC-3
[0097] Experimental steps:
[0098] A gold electrode treatment:
[0099] 1 First drop a drop of separation reagent on the gold electrode, after 15 minutes, wash it off with double distilled water.
[0100] Separation reagent: H 2 o 2 : Concentrated H 2 SO 4 =7:3 (volume ratio)
[0101] 2 with Al 2 o 3 The slurry polishes the gold electrodes. When grinding the gold electrode, the force should be uniform, and the gold electrode must be vertical.
[0102] 3 Rinse the gold electrode with double distilled water, and dry the electrode with filter paper and e...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com