Construction and screen method of novel RNA interference vector
A technology of RNA interference and screening methods, which is applied in the direction of recombinant DNA technology and the use of vectors to introduce foreign genetic materials, etc. It can solve the problems of high experimental cost, cumbersome procedures, and heavy workload, so as to reduce the base error rate and improve experimental efficiency. , the effect of reducing the cost of experiments
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] The invention is used to construct a plant artificial microRNA (amiRNA) expression vector which uses miR319a of Arabidopsis thaliana as the backbone and chalcone synthase (CHS) gene as the interference target gene.
[0039] 1) Transformation of the cloning vector
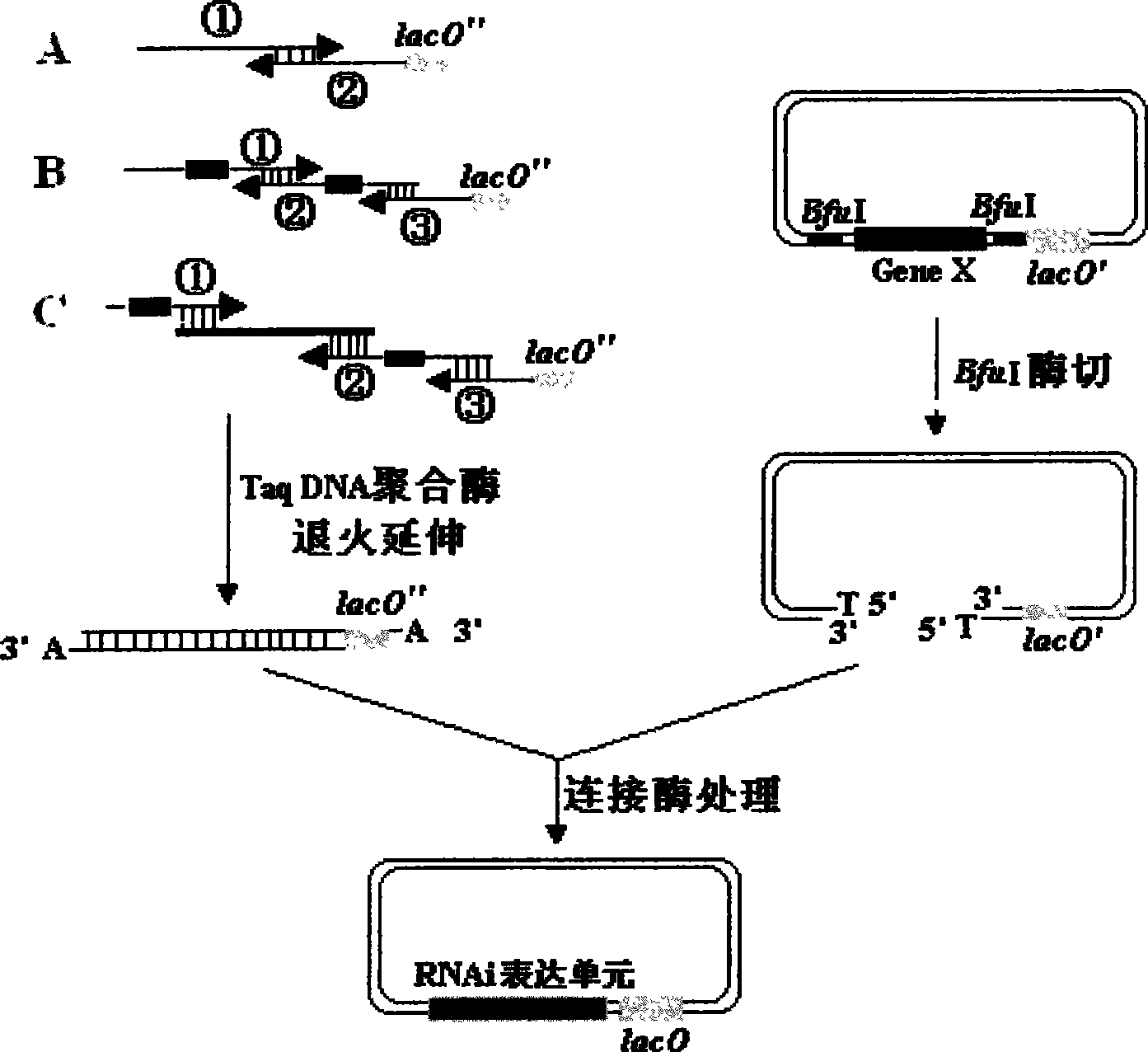
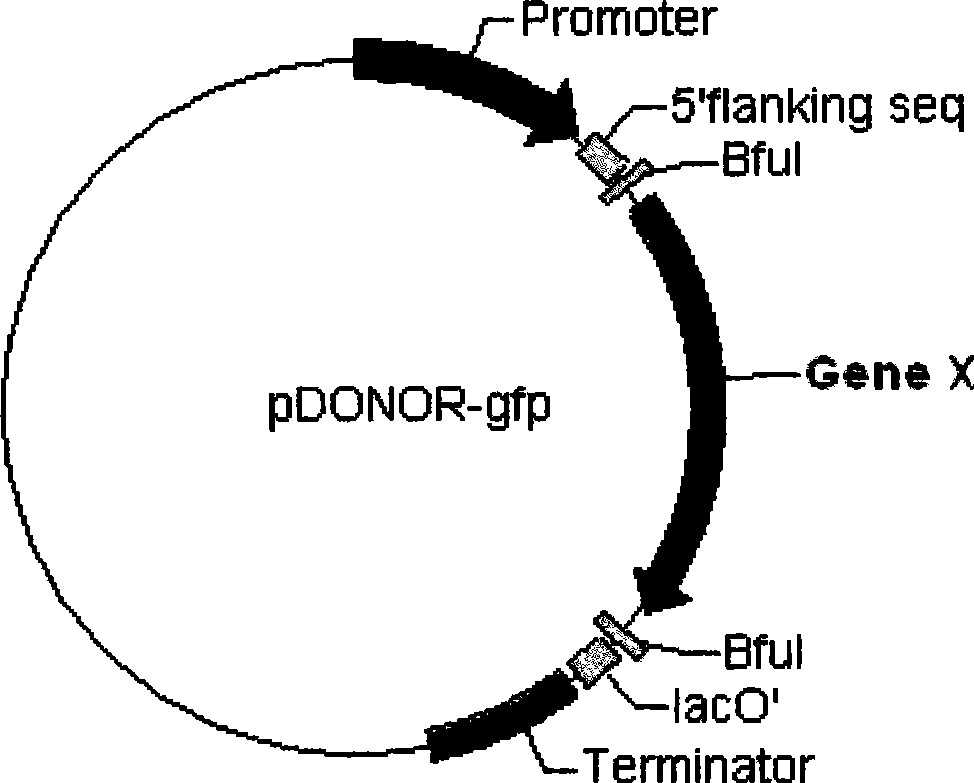
[0040] For the needs of subsequent experiments, the selected cloning vector is a donor plasmid that can be used in the mating-assisted genetically integrated cloning (MAGIC) method. After analysis, it was found that the plasmid DNA sequence carried two BfuI sites and one lacO sequence, the two BfuI sites were mutagenized into BamHI sites, and the lacO sequence was deleted and mutated. Insert the following fragments at the multiple cloning site of the transformed vector: "-promoter (P35S)-5' flanking sequence-BfuI site-gfp expression unit-BfuI site-lacO partial sequence-terminator (Tnos)- ", to obtain the transformation vector pDONOR-gfp. (See figure 2 )
[0041] 2) Formation of artificial microRNA-miR319...
Embodiment 2
[0051] The invention is used to construct an animal artificial microRNA (amiRNA) carrier with mammalian miR30 as the backbone and agtagattaccactggagtct as the interference target sequence.
[0052] 1) Transformation of the cloning vector
[0053] The vector pBluescript SK(+) sequence was analyzed, and the BfuI site and lacO sequence on the sequence were mutagenized. Insert the following fragments at the multiple cloning site of the transformed vector: "-promoter (CMV)-5' flanking sequence-BfuI site-gfp expression unit-BfuI site-lacO partial sequence-terminator (SV40PolyA)- ".
[0054] 2) Formation of artificial microRNA—mir30-X containing interference target sequence
[0055] According to the principles in step 2) of the specific method of the present invention, analyze the miR30 sequence, design and synthesize a universal primer pUR and a pair of specific primers pF & pR, mix the 3 primers, anneal at 25°C with Taq DNA polymerase, 72 °C extension, the obtained product is mi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com