Gene chip, kit and method for detecting common pathogenic bacteria of piglets
A gene chip and pathogenic bacteria technology, applied in the field of gene chip detection of common pathogenic bacteria in piglets, can solve the problems affecting the quality of growth and development, incomplete function of physiological system, low immunity, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0105] Example 1. Design of gene chip-specific probes
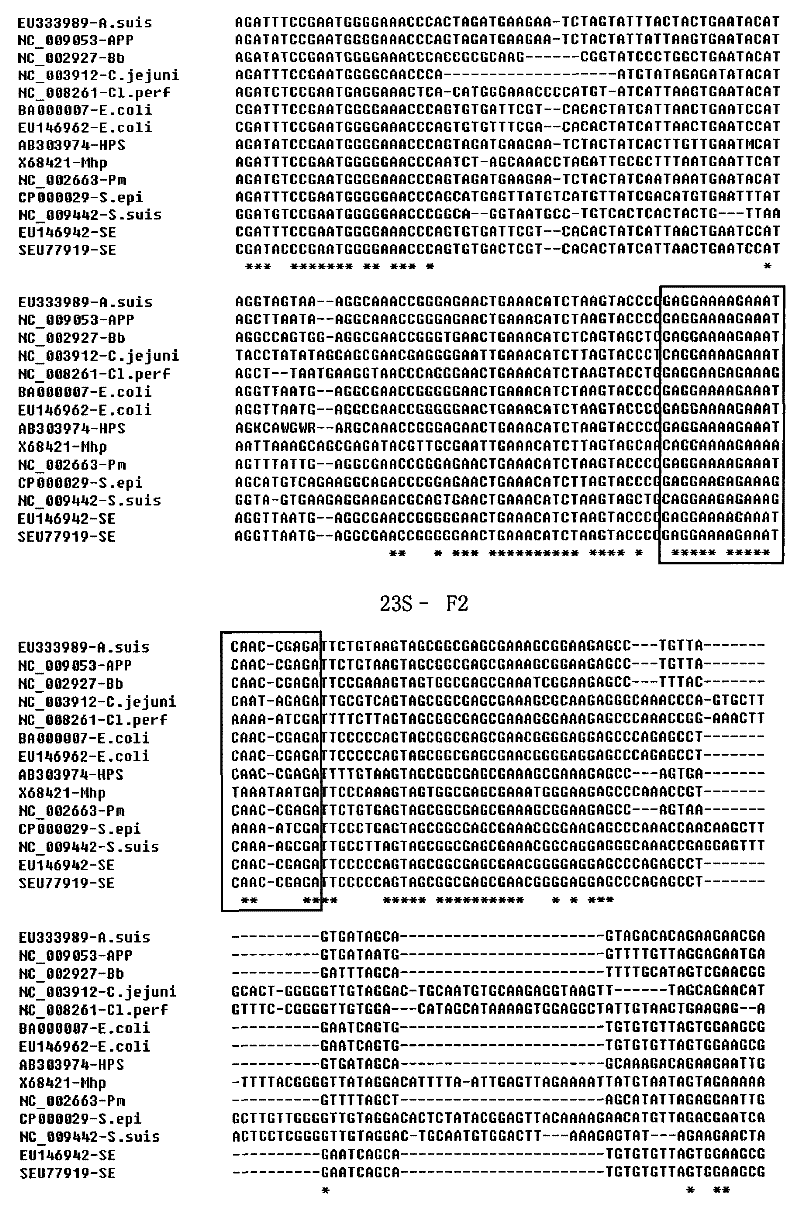
[0106] Step 1: 23S rRNA sequence alignment analysis of target bacteria
[0107] Collected 108 23S rRNA complete or nearly complete gene sequences of 12 target pathogenic bacteria from Genbank (there are too few available sequences for individual bacteria, our laboratory sequenced 6 and submitted them to Genbank. A. suis has no choice but to submit them to us a sequence). The 108 sequences were constructed by species, compared with the clustalW program of DNAStar software, and 1 or 2 representative sequences of each bacterium were selected (14 in total), among which the representative sequence of Staphylococcus epidermidis (S.epi) was registered The number is CP000029, Haemophilus parasuis (Hps) is AB303974, Actinobacillus pleuropneumoniae (App) is NC009053, Pasteurella multocida (Pm) is NC_002663, Actinobacillus suis (A.suis) is EU333989, Bordetella bronchiseptica (Bb) is NC_002927, Clostridium perfringens (Clp) is NC_0...
Embodiment 2
[0127] Embodiment 2. Primer generality test
[0128] Step 1. Extract DNA
[0129] Take appropriate cultures of the 12 target bacteria in Table 1 (i.e., the reference strains with asterisks in Table 1), and use the Tiangen Bacterial Genome Kit to extract bacterial genomic DNA. The purity and concentration of DNA were measured by ultraviolet spectrophotometer, and 1ng / μl DNA was used for PCR.
[0130] Or obtain crude DNA by boiling: take the bacterial suspension, centrifuge at 12,000rpm for 2 minutes, pour off the supernatant or pick colonies, add 100-400μl double distilled water to shake and suspend, OD600 is 1.0-2.0, put it in a boiling water bath and boil Or heat in a metal bath at 95-100°C for 10-15 minutes, ice-bath for 10 minutes, centrifuge at 12,000 rpm for 2 minutes, and take 2 μl of the supernatant as a PCR template.
[0131] Use each pair of universal primers designed in Example 1 to perform conventional single-plex PCR on the DNA of 12 pathogenic bacteria respectiv...
Embodiment 3
[0133] Example 3. Optimizing the amplification system
[0134] The purpose of asymmetric PCR is to obtain a large number of single-stranded PCR products by adding more fluorescent-labeled downstream universal primers and less non-fluorescent upstream universal primers, and increase the hybridization efficiency of PCR products and single-stranded probes.
[0135] Each pair of universal primers was optimized three times using the genomic DNA of two pathogenic bacteria as templates. In the PCR reaction system, the final concentration of the downstream universal primer was fixed at 1 μM, and serially diluted upstream universal primers were added to make the final concentration decrease (1 μM, 0.2 μM, 0.1 μM, 0.05 μM, etc.), that is, the ratio of the upstream and downstream universal primers was 1:1, 1:5, 1:10...1:60.
[0136]The optimized results of two pairs of universal primers are
[0137] 23S-F1:23S-R1 is 1:10, 23S-F2:13S-R2 is 1:30. The ratio between two pairs of universal...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com