Gene series technology for increasing main component content of gene engineering isovaleryl selectomycin
A technology for isovalerylspiramycin and spiramycin is applied in the field of gene tandem technology for improving the content of main components of genetic engineering isovalerylspiramycin, and achieves the effects of improving control, simplifying extraction process and reducing production cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
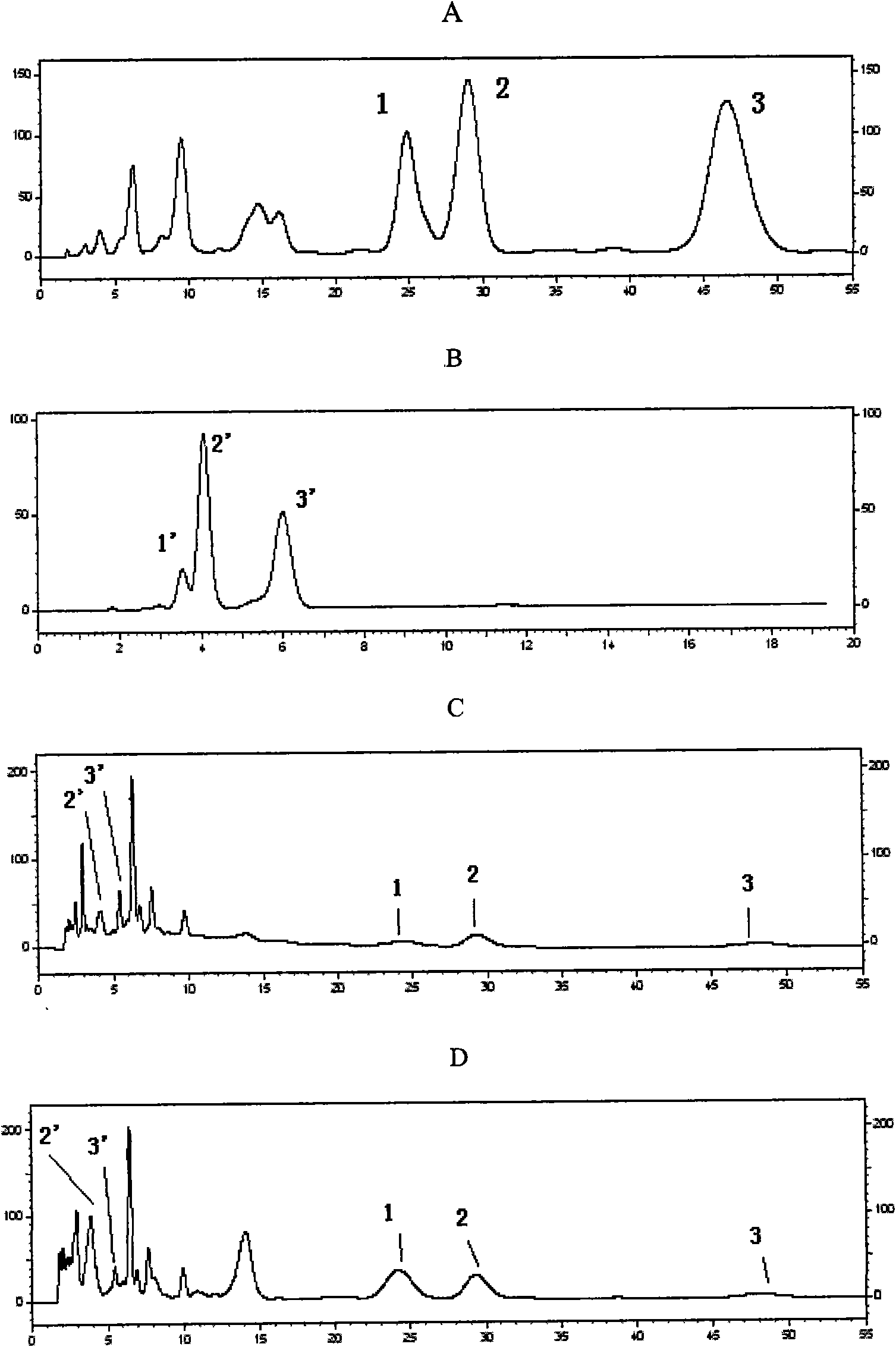
Image
Examples
Embodiment 1
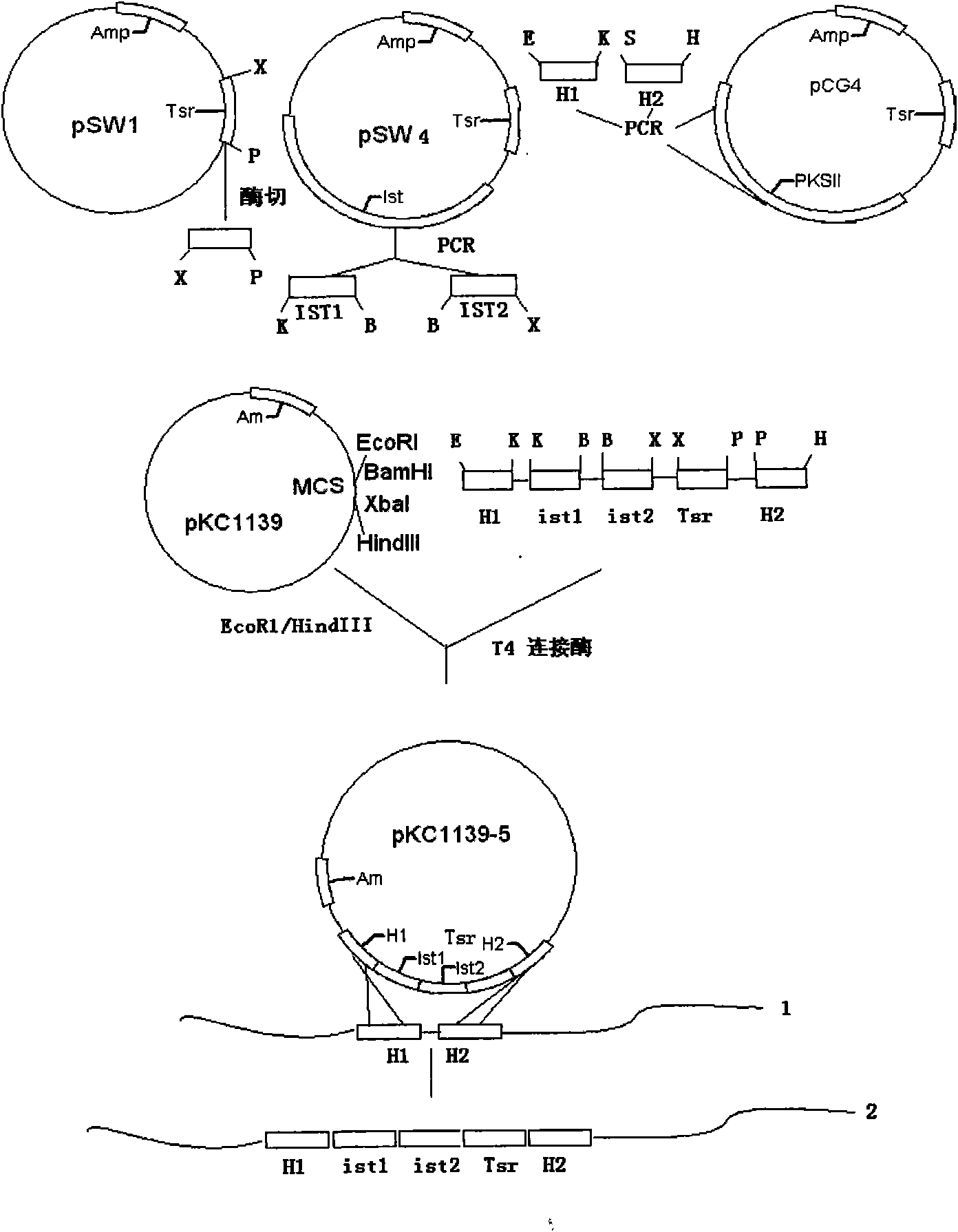
[0039] : The concatenation of two double-copy ist genes with their own promoters
[0040] For the tandem of double-copy ist genes 1 and 2, use the repeated tandem method including its own promoter, and design P1-P2 and P3-P4 primers at the ist gene start codon ATG upstream - 129kb and terminator TAG downstream - 41 (Table 1), using the pSW4 plasmid containing the ist gene as a template, two genes of about 1.3 kb with different restriction sites were amplified by PCR, namely the ist1 gene (KpnI-BamH I) and the ist2 gene (BamHI- Xba I) fragment, the same BamH I restriction site can be used to carry out the tandem connection between the ist gene with its own promoter in two copies.
Embodiment 2
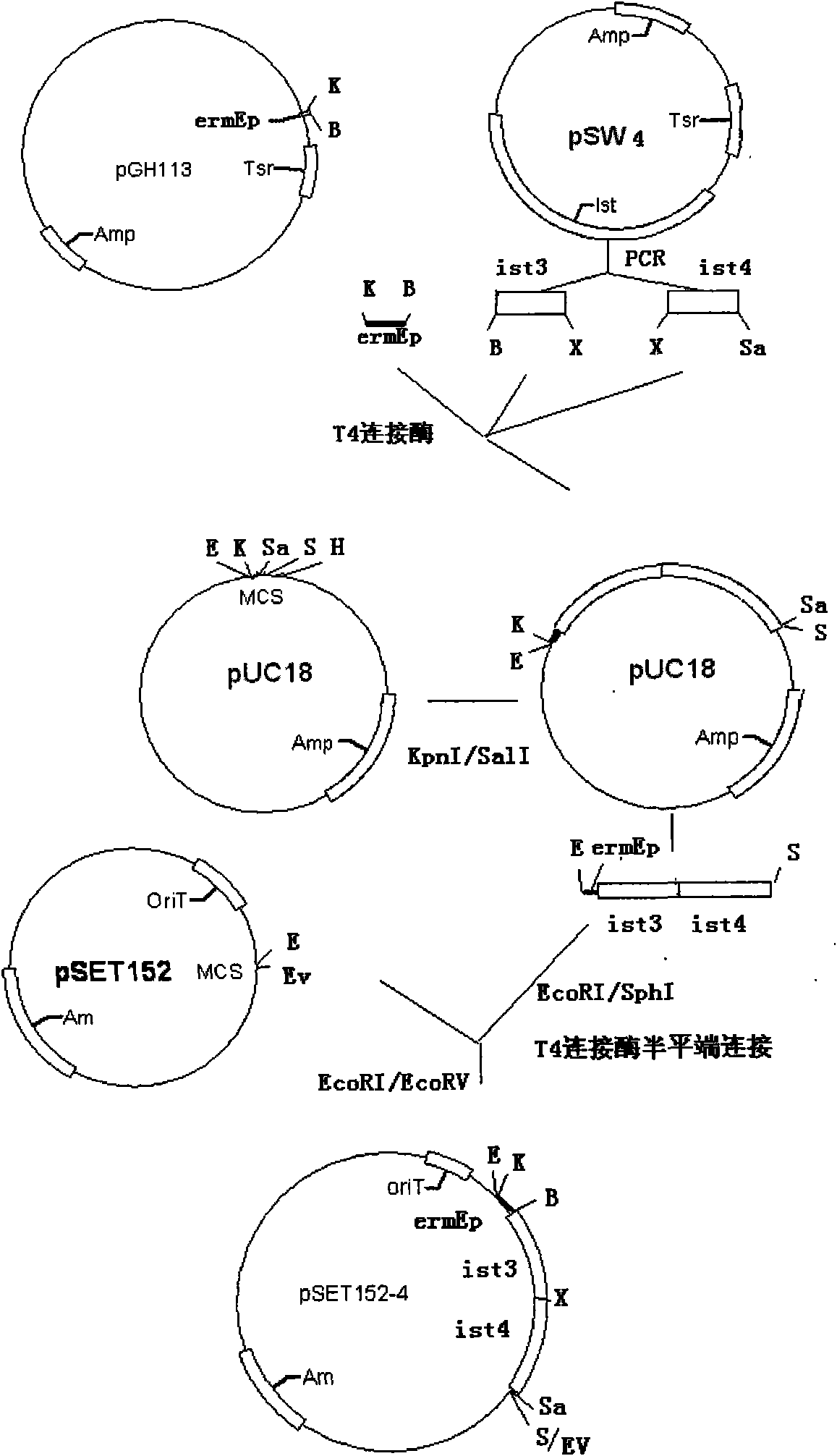
[0041] : Construction of double-copy ist gene tandem transcription
[0042] Design P5-P6 primers at the ist gene start codon ATG upstream -111bp and terminator TAG downstream -24bp (excluding the transcription termination sequence), and design P7 at the ATG upstream -105bp and terminator downstream -80bp (including the transcription termination sequence) -P8 primer (Table 1), double copy ist gene tandem primer design site see figure 1 . Using the pSW4 plasmid containing the ist gene as a template, the ist 3 gene (containing BamHI-XbaI site) and ist 4 gene (containing XbaI-SalI site) fragments were respectively obtained by PCR, and the same XbaI restriction site was used to perform double A tandem of copies of the ist gene.
Embodiment 3
[0043] : construction of homologous recombination double exchange ist gene double copy plasmid (pKC1139-5)
[0044] Using P9-P10 primers to pCG4 plasmid as a template, the homologous fragment H of about 1.1 kb was amplified by PCR 1 , using P11-P12 primers to pCG4 plasmid as a template, the homologous fragment H of about 1.1kb was amplified by PCR 2 , used to construct fragments of homologous recombination double crossover on both sides of the ist gene. Digest the pWS1 plasmid with Xba I-Pst I [Shang Guandong et al The J of antibiotics 200154 (1): 66-73] to obtain a 1.3kb thiostrepton resistance gene (Tsr r ) fragment as a screening marker for homologous recombination double crossover. H 1 Fragment (EcoR I-Kpn I), ist1 gene (Kpn I-BamH I), ist 2 gene (BamHI-Xba I) described in Example 1, and Tsr r Genes (XbaI-PstI) and H 2 The (PstI-Hind III) fragment was directly connected to the thermosensitive plasmid vector pKC1139 plasmid [Bierman M, et al.Gene, 1992, 116: 43-49] on ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com