Polynucleotide related to mitochondrial membrane potential decrease and coded polypeptide and application thereof
A technology of mitochondrial membrane potential and polynucleotides, applied in the field of polypeptides
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0073] Example 1. Amplification of target gene by two-step flux RT-PCR technology
[0074] (1) Search the NCBI refseq database for unknown human function prediction genes to obtain the unknown human function gene sequence, and use the Human_est database to perform sequence correction by the BLASTn method. The final sequence is set as the following sequence: SEQ ID NO:1 , SEQ ID NO: 3, SEQ ID NO: 5, SEQ ID NO: 7. Design specific primers for genes MMPRG1, 2, 3, 4 based on such sequences:
[0075]
[0076] (2) Using the above primers, select templates from the existing cDNA library and tumor library according to the expression profile of the target gene, and perform initial amplification. The current library includes 12 normal human tissues (heart, pancreas, testis, ovary, prostate, colon, small intestine, skeletal muscle, thymus, lymph nodes, tonsils, white blood cells); 6 human tumor tissues (lung cancer, pancreatic cancer, ovarian cancer) , Prostate cancer, colon cancer, breast c...
Embodiment 2
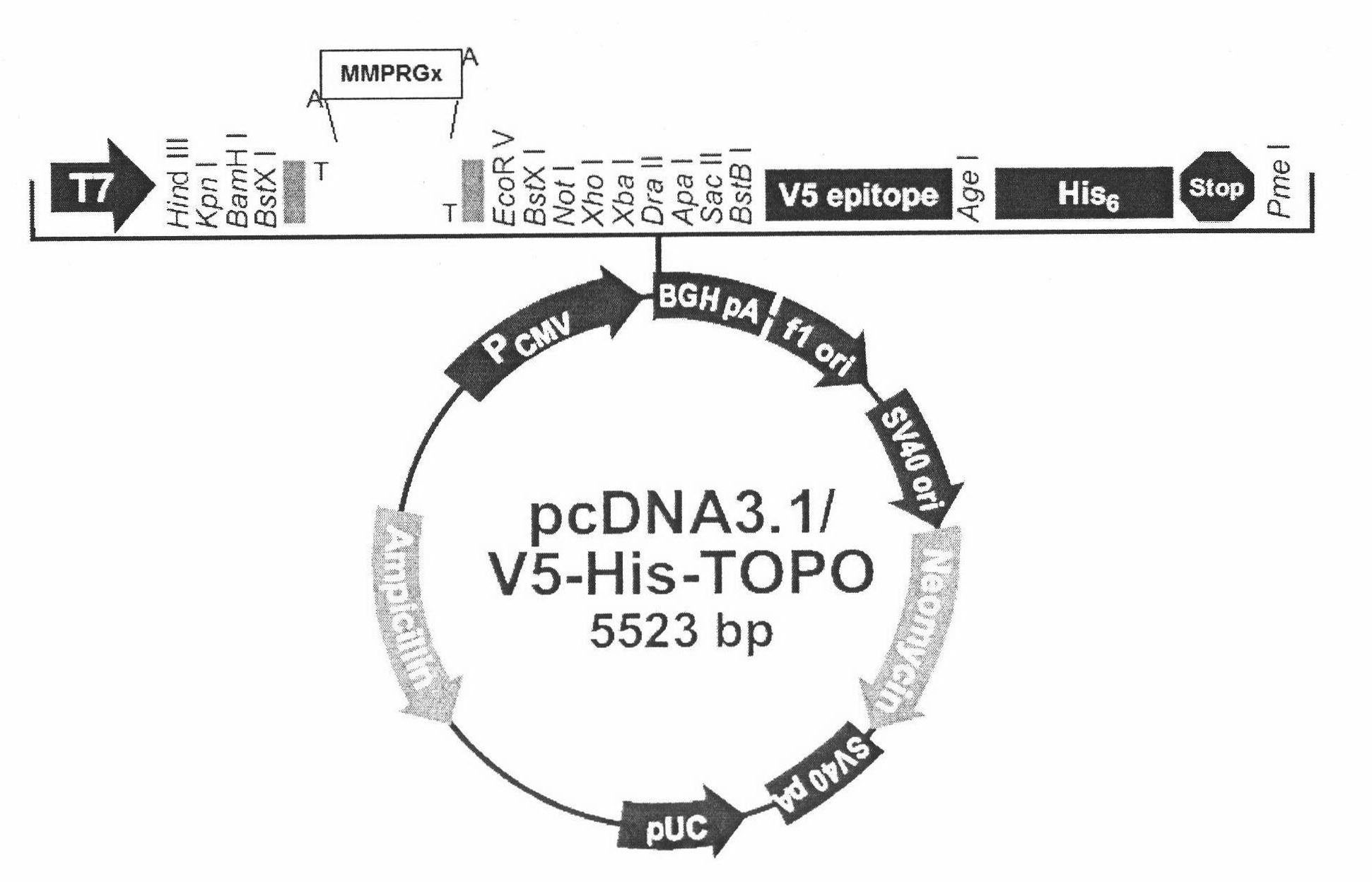
[0088] Example 2. Construction of eukaryotic expression vector of target gene
[0089] The two-expanded purified product and the eukaryotic expression vector pcDNA3.1 / V5-His-TOPO (Invitrogen, abbreviated as pcDT) were ligated according to the conditions recommended by the kit manufacturer. The ligation product was transformed into E. coli DH5α by electroporation. The transformant was grown on a solid LB plate medium containing ampicillin. The single cloned colony was selected and the plasmid was extracted. The digested product was digested with EcoRI. The digested product was identified by agarose gel electrophoresis. Select positive clones with inserts, and select the correct forward insert clones by sequencing (ABI PRISM 3700 DNA Analyzer), and name them MMPRG1, 2, 3, and 4 respectively.
[0090] At the same time, the culture fluid was collected, and the precipitated protein was analyzed by SDS-PAGE to obtain MMPRG1, 2, 3, 4 polypeptide.
[0091] MMPRG1, 2, 3, 4 protein analysis r...
Embodiment 3
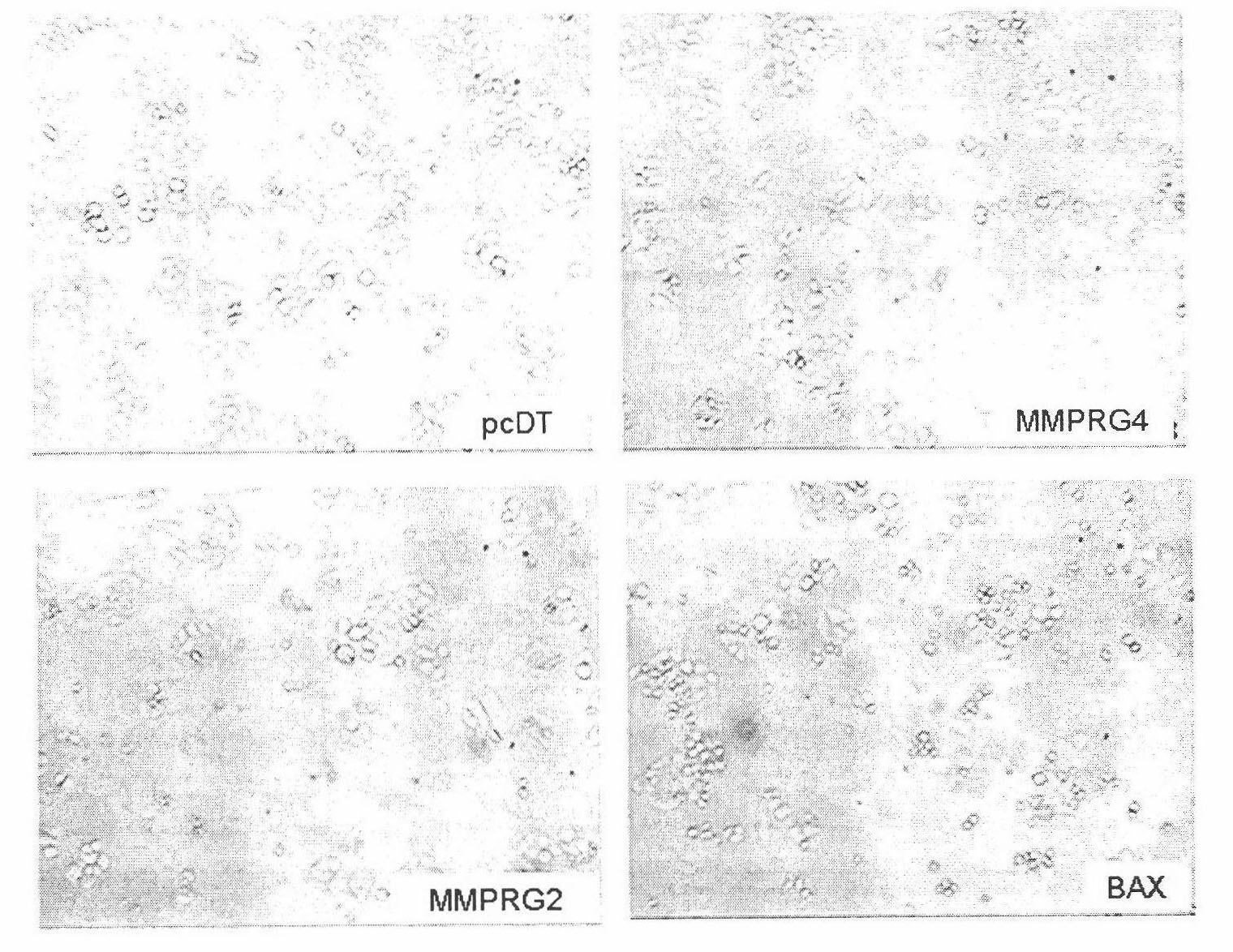
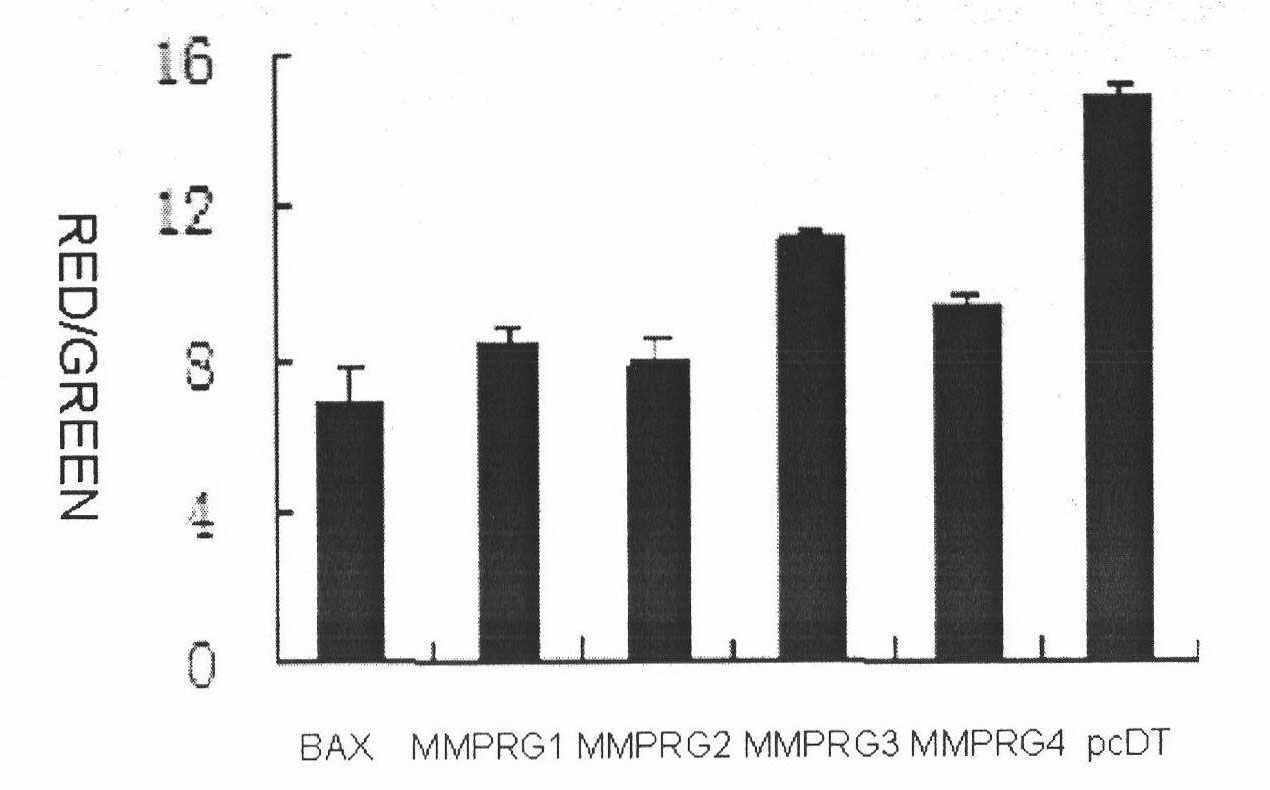
[0092] Example 3. Using JC-1 dye to determine the effect of the target gene on the decrease of mitochondrial membrane potential
[0093] After transfecting Hela cells with the target gene, use JC-1 dye to stain the cells, operate as described in the product manual, and observe the changes in cell morphology under a fluorescence microscope. The specific steps are as follows:
[0094] In this experiment, a 96-well cell culture plate was used, and each gene to be tested was set up with 3 parallel repetitive wells, and pcDT empty plasmid and Bax plasmid were used as empty vector and positive control respectively. The dosages in the following steps are all single-hole dosages.
[0095] (1) Cell culture: add 1.2×10 4 Two Hela cells (ATCC Number: CRL-11268) were plated on 96-well cell culture plates (Costar, 3599) with DMEM (Dulbecco's modified Eagle's medium) medium (Hyclone, SH0022.02) containing 10% fetal bovine serum ) (100μl culture medium / well), set at 37℃, 5% CO 2 Culture in a cell...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com