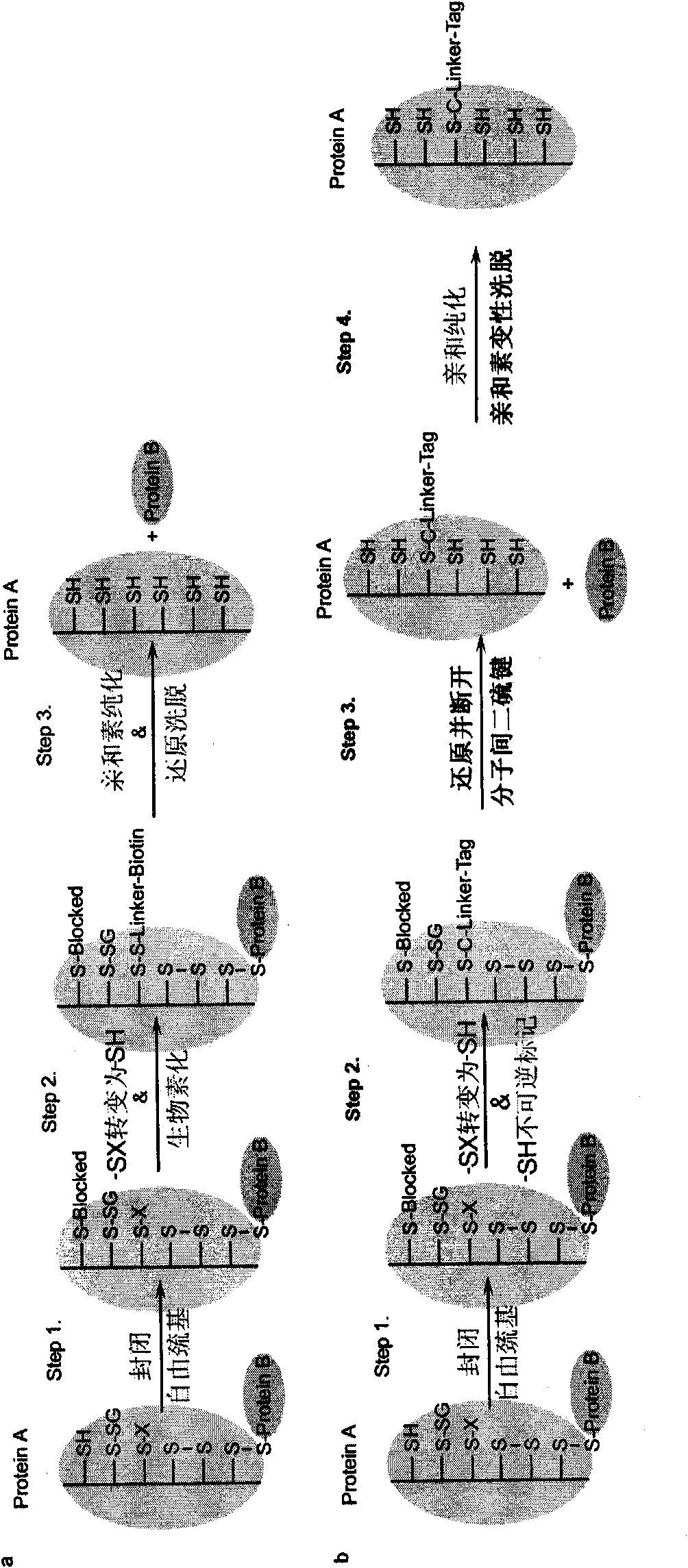
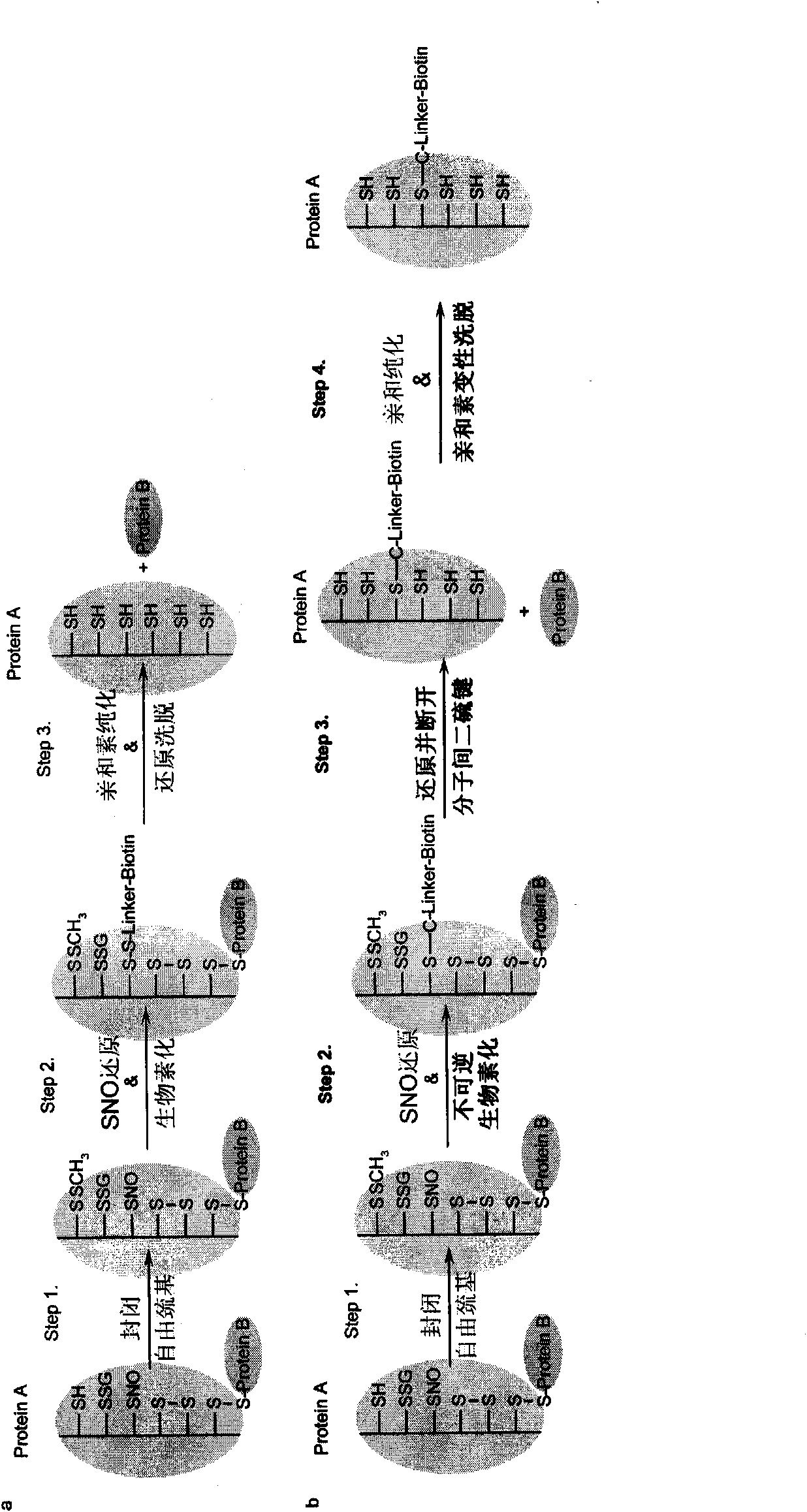
Specific detection method of protein or polypeptide cysteine sulfydryl modification and application thereof
A technology of cysteine sulfhydryl and cystine sulfhydryl, which is applied in the field of detection of post-translational modification of proteins or polypeptides, and can solve problems such as interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] Example 1: Materials and methods
[0107] animal
[0108] We used 4-5 week old C57BL / 6 mice (SPF grade, Beijing Weitong Lihua Experimental Animal Technology Co., Ltd.). All animal-related procedures were approved by the Animal Care Committee of the Institute of Biophysics, Chinese Academy of Sciences.
[0109] Raw Biotin Conversion Method
[0110]Mouse brain was homogenized using 5ml of HEN buffer (25mM HEPES (4-(2-hydroxyerhyl)piperazine-1-erhanesulfonic acid, 4-hydroxyethylpiperazineethanesulfonic acid) pH 7.7, 0.1mM EDTA (ethylenediamine Tetraacetic acid), 10mM neocuproine (2,9-dimethyl-1,10-phenanthroline)), add 1% NP40 (Nonidet P-40, ethylphenyl polyethylene glycol) in the homogenate, protease inhibition reagent, 1 mM PMSF (phenylmethylsulfonyl fluoride), centrifuged at 12,000 g at 4°C for 10 minutes. Half of the supernatant was treated with 40 μM NO donor GSNO (S-nitrosoglutathione) for 30 minutes at room temperature. The NO donor was removed by acetone preci...
Embodiment 2
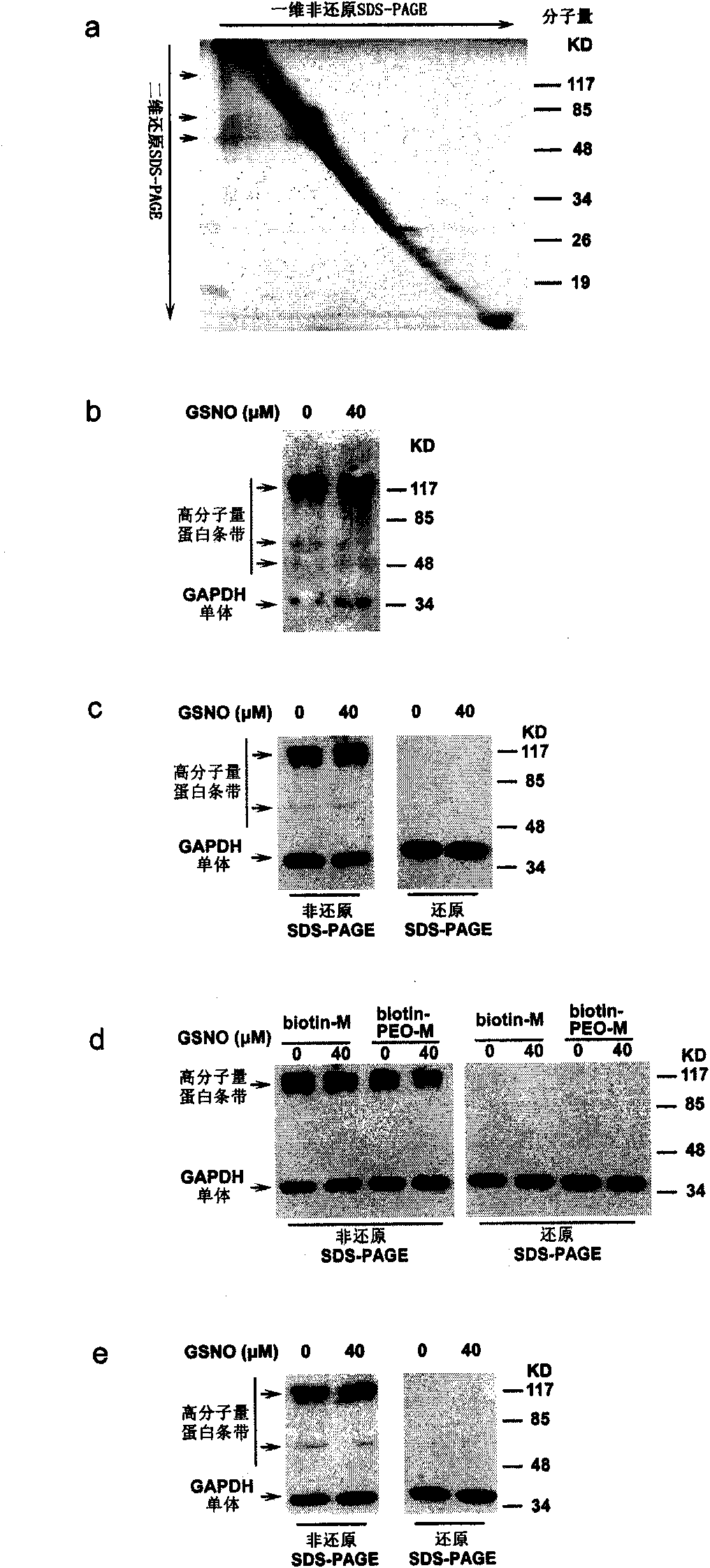
[0117] Example 2 uses "diagonal eleetrophoresis" 10,11 and Western blot analysis to detect the original biotin conversion method is interfered by intermolecular disulfide bonds ( image 3 )
[0118] After the first-dimension non-reducing SDS-PAGE, the entire electrophoresis lane was excised for the second-dimension reducing SDS-PAGE. If there is no intermolecular disulfide bond, the molecular weight of the protein band will not change, and it will be on a diagonal; if there is an intermolecular disulfide bond, then the molecular weight of the protein will decrease and move below the diagonal 10,11 . When eluting protein samples that have undergone the biotin conversion method, we use a non-reducing denaturing elution method: high temperature and high concentration of denaturant. This ensures that after elution, potential intermolecular disulfide bonds remain intact and can be analyzed by subsequent diagonal electrophoresis.
[0119] We treated mouse brain homogenates with...
Embodiment 3
[0121] Example 3 uses different sulfhydryl blocking reagents, purification reagents, and biotinylation reagents. Intermolecular disulfide bonds exist in all cases ( Figure 4 )
[0122] We doubted whether these intermolecular disulfide bonds were formed during the MMTS-blocking step, so we replaced the reversible thiol-blocking reagent MMTS with an irreversible thiol-blocking reagent, NEM (N-ehylmaleimide), but we Intermolecular disulfide bonds were still found in subsequent protein samples ( Figure 4 a). Then we replaced the biotinylation reagent biotin-HPDP with biotin-maleimide or the purification reagent streptavidin sepharose beads with avidin sepharose beads, the intermolecular disulfide bond still exists ( Figure 4 b, Figure 4 c). These results imply that the detected intermolecular disulfide bonds are not derived from the biotin conversion process, but from the original sample of mouse brain homogenate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com