Sequencing of nucleic acids
A nucleic acid and nucleic acid base sequence technology, applied in the field of high-throughput parallel DNA sequencing, can solve the problem of large differences in the copy number of captured sequences, and achieve the effects of reducing sequencing redundancy requirements, reducing sequencing time, and high capacity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
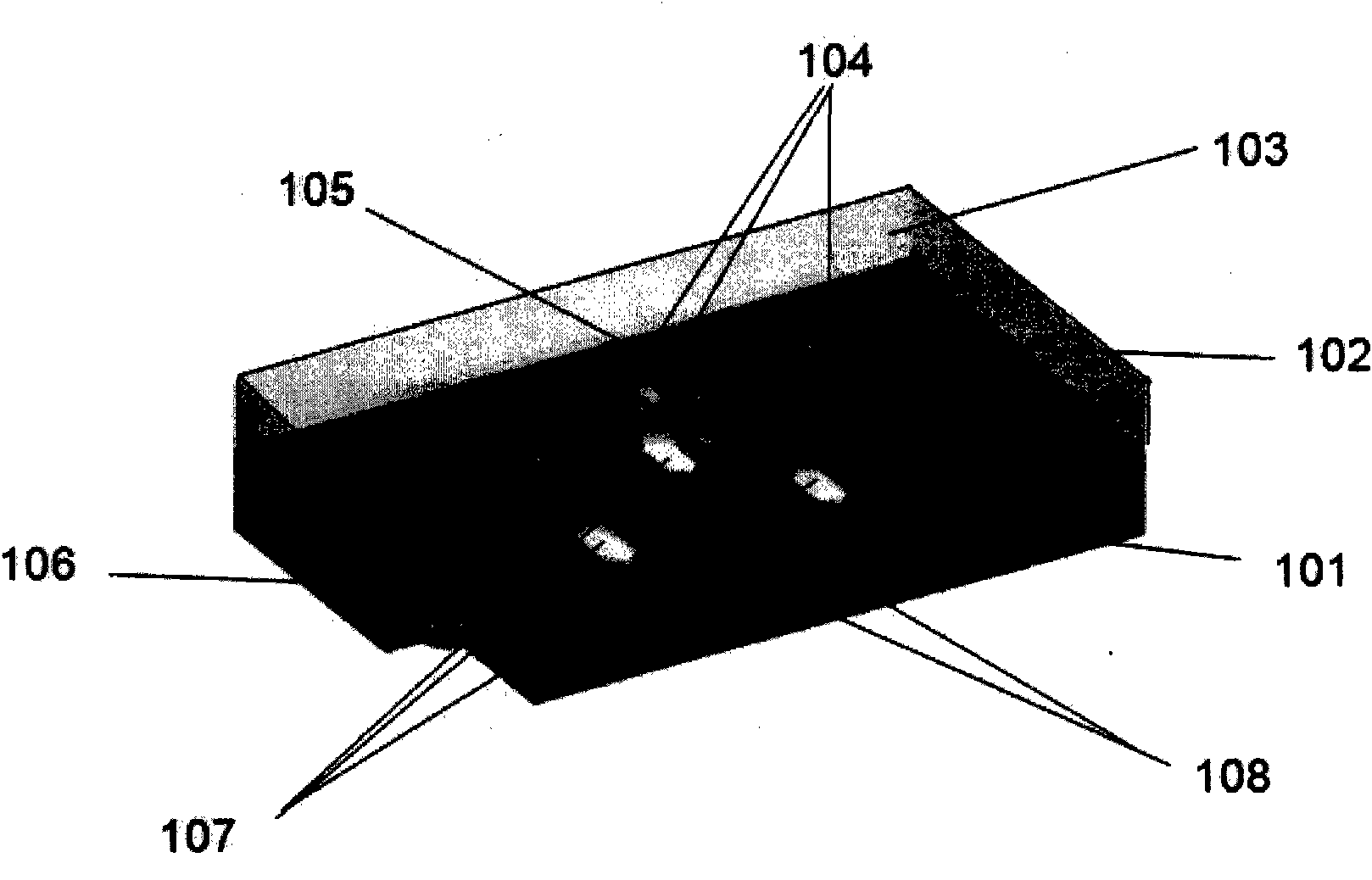
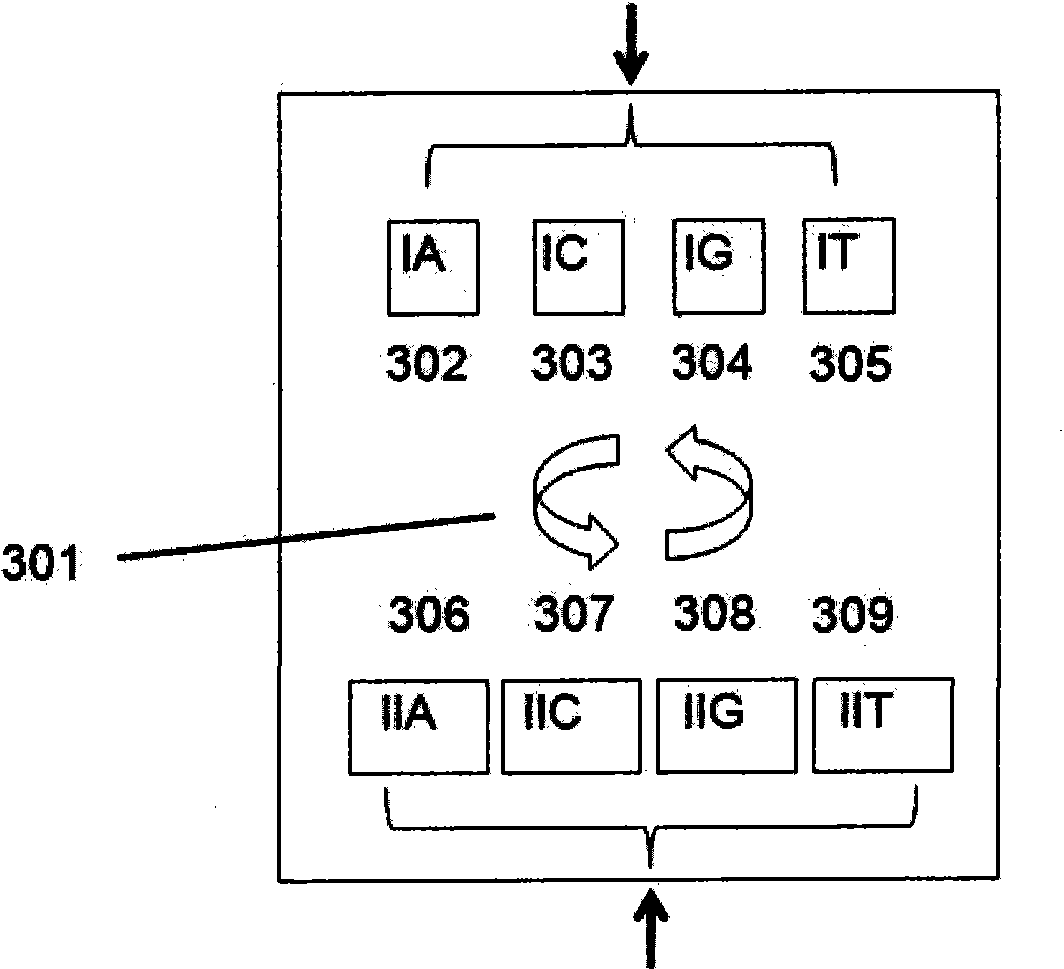
[0058] The methods of the invention are not limited to the types of molecules already discussed. In a preferred embodiment of the present invention, DNA, RNA, polypeptides, carbohydrates or other molecules can be synthesized in situ on addressable nanobeads in an amendable manner. At the same time, the synthesis method of the present invention is not limited to the number of reaction chambers that can be used to synthesize molecular nanobeads. Such as Figure 5 As shown, when a single reaction chamber is used, it is conceivable that each monomer species is added to multiple reaction chambers. Reaction chambers can also be utilized simultaneously far beyond a single step. Additional synthetic steps include the utilization of dimers and trimers or longer available elements.
[0059] The number of different elements to be added will determine the minimum number of reaction chambers necessary, such that one reaction chamber is necessary for each element. For example, using nat...
Embodiment 1
[0119] Example 1 Sequence in Elution Capillary Bundle
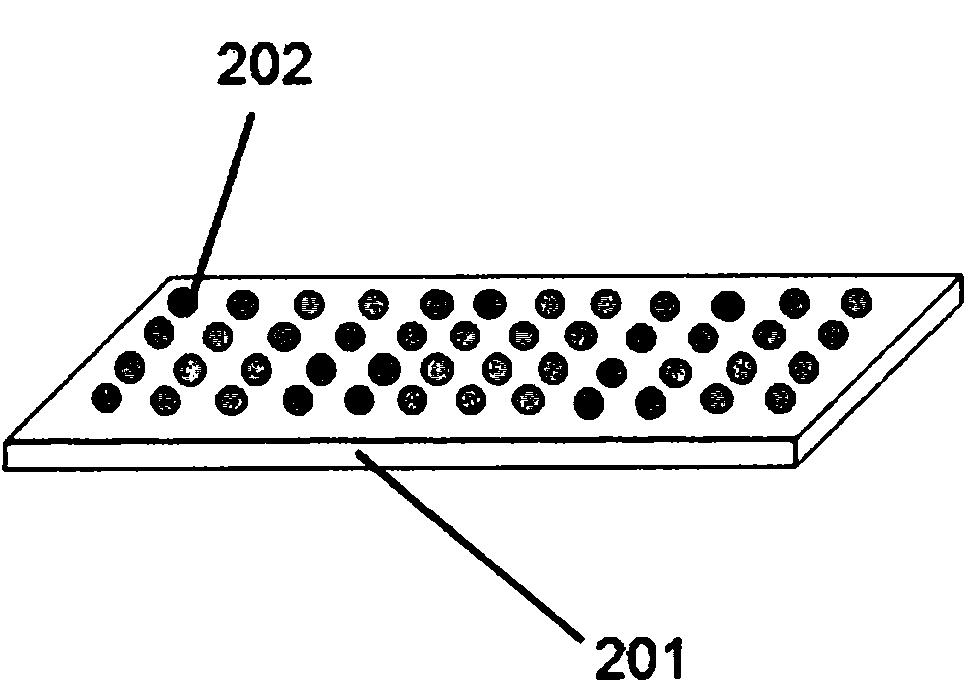
[0120] The sequencing CE block is made of drawn glass, and a capillary with an inner diameter of 100 m is used to form a depressed channel bundle with a channel cross-sectional area of 2x3 mm2 and a length of 5 cm. Fill the sequencing channel with 10% PAGE gel by capillary effect, and allow the solution to flow through half of the bottom area (perpendicular to the channel) to load the sample (see below). The samples used contained oligonucleotides of different lengths labeled with four fluorescent dyes. These 4 oligonucleotides are FAM-18mer, Cy3-6mer, Cy3-38mer and FAM-46mer. Afterwards, place the sequencing CE block in the horizontal electrophoresis device for a certain period of time (minutes), take it out and use a fluorescence microscope (Olympus BX41EPI fluorescence research microscope) to obtain images of the outer end surface, and then put it back into the electrophoresis device to continue. Repeating this pro...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
| height | aaaaa | aaaaa |
| length | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com