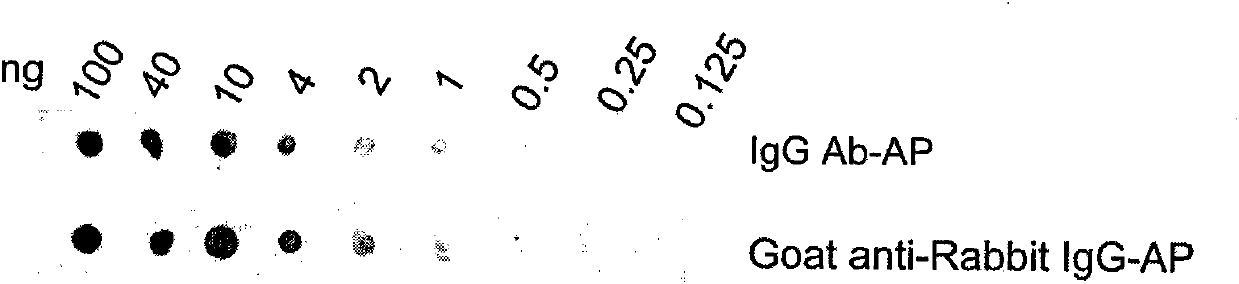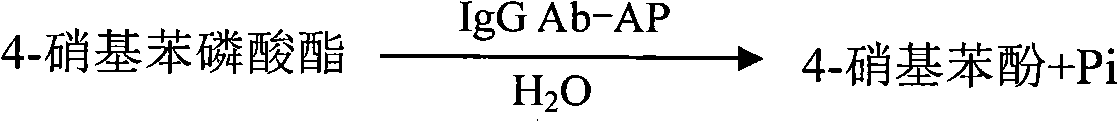IgG antibody affinity peptide-alkaline phosphatase fusion protein
A technology of fusion protein and phosphatase, which is applied in the field of bioengineering, can solve problems such as fusion label blockage, and achieve good effect, convenient use and high activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 Construction of IgG antibody affinity peptide-alkaline phosphatase fusion protein gene expression vector
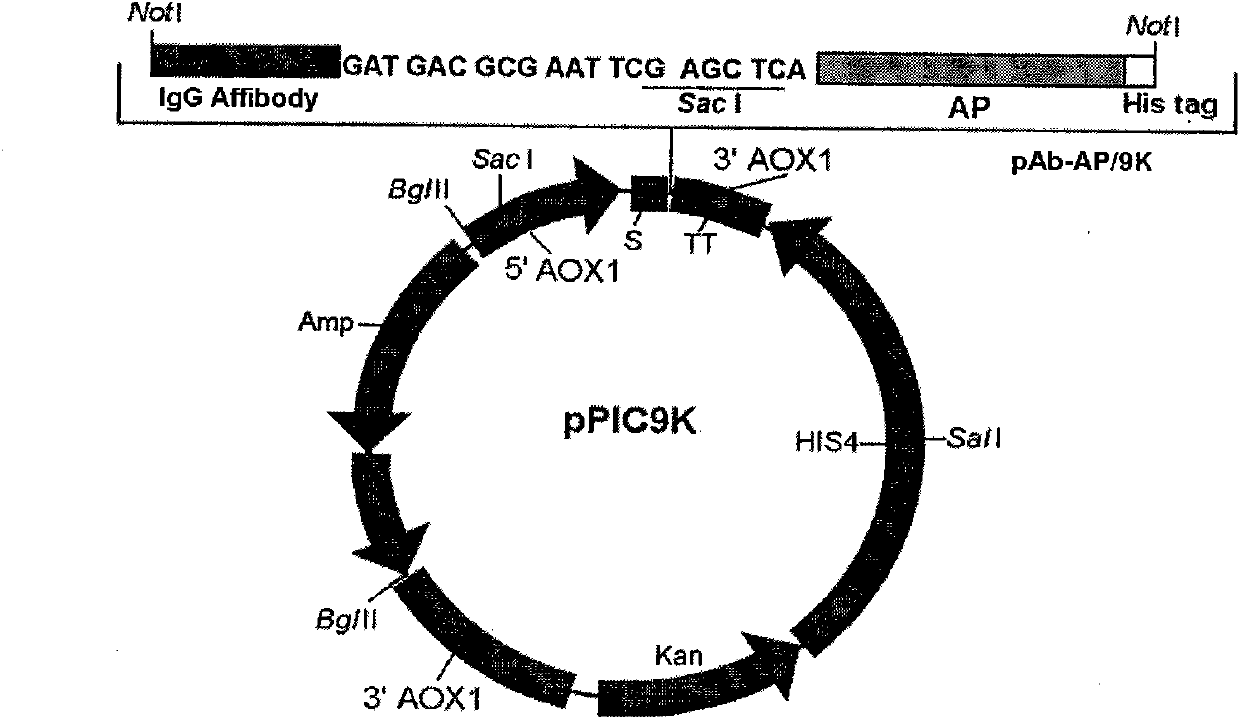
[0060] In order to link the alkaline phosphatase gene with the IgG affinity peptide gene, first provide restriction endonuclease cleavage sites upstream and downstream of the secreted alkaline phosphatase gene (secreted alkaline phosphatase, SEAP, GenBank Accession No: U35316) Spot Sac I and Pst I, amplify the alkaline phosphatase gene (1467bp) without N-signal peptide and C-terminal extended peptide by PCR, insert IgG affinity peptide downstream (Sac I and Pst I sites of pEZZ18) ; The second step provides a restriction endonuclease cutting site Not I at the upstream and downstream of the IgG affinity peptide-AP gene, and is amplified by PCR and inserted into the Not I site of pPIC9K to construct the Pichia pastoris expression vector pIgG Ab-AP / 9k, such as figure 1 shown.
Embodiment 2
[0061] Example 2 Electrotransformation of Pichia pastoris GS115
[0062] For transformation of pAb-AP / 9k in Pichia pastoris GS115 and subsequent integration into the genome, the vector was first linearized with Sal I and electrotransformed using a Gene Pulser Xcell electroporator:
[0063] 1) Pick a single colony of yeast, inoculate in 5mLYPD medium, and culture overnight at 30°C and 250rpm;
[0064] 2) Inoculate 100-500 μL of the overnight culture into 500 mL of LYPD medium, culture at 30°C, 250 rpm to OD 600 Reach 1.3-1.5;
[0065] 3) Centrifuge the bacterial solution at 4°C, 3000 rpm for 5 minutes, and resuspend the bacterial cells with 500 mL of ice-cold sterile water;
[0066] 4) Centrifuge according to step 3, and resuspend the bacterial cell pellet with 250 mL ice-cold sterile water;
[0067] 5) Centrifuge according to step 3, and resuspend the bacterial cell pellet with 20 mL ice-cold 1M sorbitol;
[0068] 6) Centrifuge according to step 3, and resuspend the cells ...
Embodiment 3
[0073] The screening of embodiment 3 high-copy bacterial strains and the expression of target protein
[0074] In order to obtain high-copy genetically engineered bacteria, selection pressure was used for screening. The correct single colony will be verified and seeded on the YPD plate containing different G418 concentrations with a gun tip. When spotting, spot the plates in order of G418 concentration from low to high, and culture at 30°C.
[0075] Inoculate high-copy single colonies that have passed G418 (4.0mg / mL) pressure screening in 25mL BMGY, and cultivate them to OD at 30°C and 280rpm 600 Reach 2-6. Collect cells by centrifugation at 3000g for 5min. Discard the supernatant, resuspend the cells with BMMY medium, transfer to 50mL BMMY, induce protein expression at 30°C, 280rpm; 1mL is sampled every 24h, and methanol is added once to reach a methanol concentration of 0.5%, and the induction ends after 120 hours.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com