Maize water-logging tolerance-related transcription factor gene zm-bRLZ, molecular marker and application
A technology of transcription factor and water tolerance, applied in the field of plant biology, can solve the problems of enhancing long-term adaptive response of Arabidopsis thaliana and the premature death of transformed plants.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Obtaining zm-bRLZ by cDNA chip screening
[0027] (1) The differentially expressed gene zm-bRLZ was obtained by microarray screening
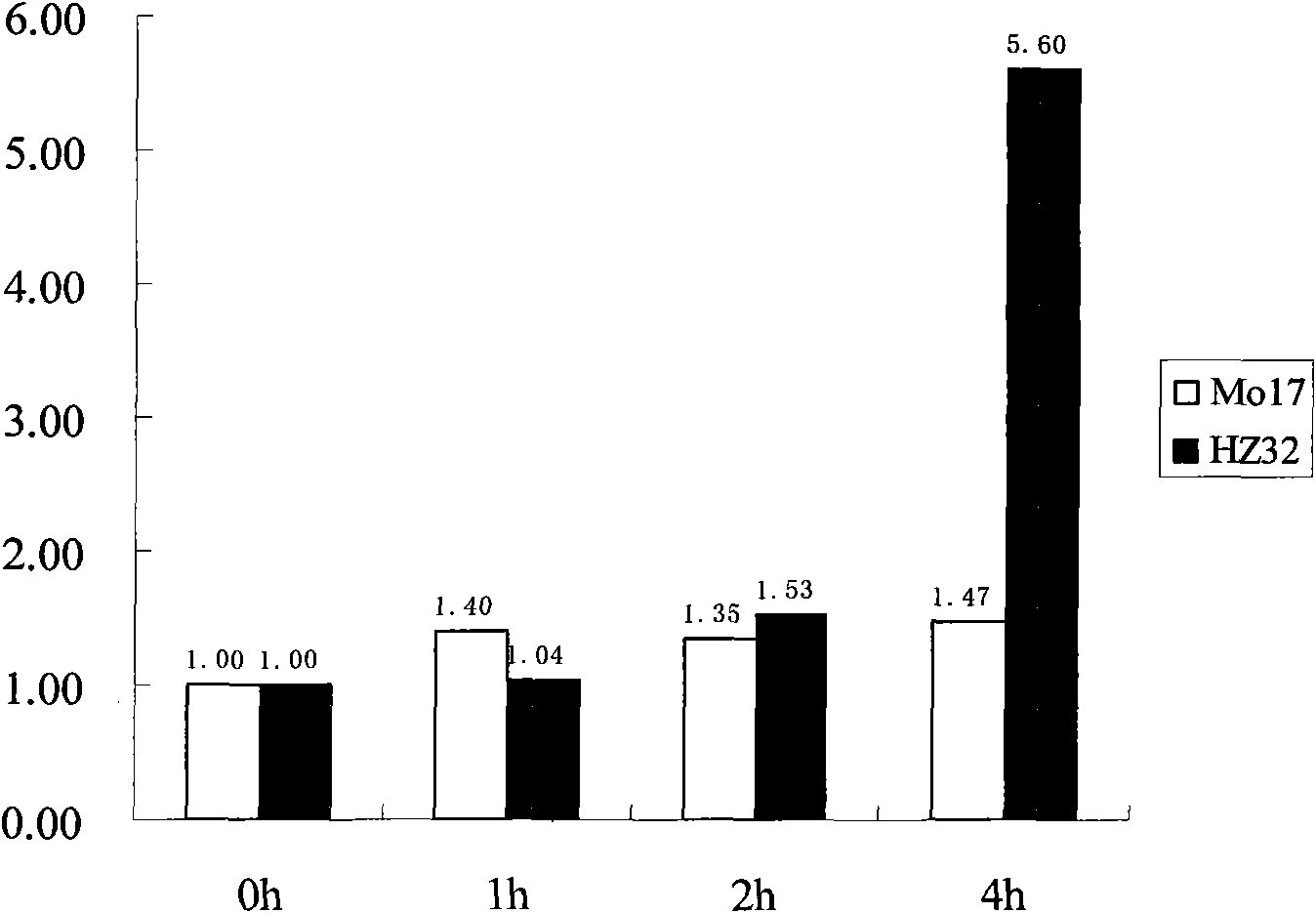
[0028] In the present invention, two corn inbred lines, Mo17 with poor water resistance and Hz32 with strong water resistance, are selected as test materials. After the seeds germinate, they are potted in sand pots until the stage of two leaves and one heart. A number of individual plants with the same growth were selected and divided into two groups on average. One group was used as a control and grown under normal water and management conditions; the other group was grown in 1× complete culture solution [component: Ca(NO 3 )2 820.7mg / L, KNO 3 505.6mg / L, MgSO 4 ·7H 2 O 616.2mg / L, KH 2 PO 4 272.2mg / L, Fe-EDTA 13.02mg / L, H 3 BO 3 2.860mg / L, MnSO 4 1.015mg / L, CuSO 4 ·5H 2 O 0.079mg / L, ZnSO 4 ·7H 2 O 0.220mg / L, H 2 MoO 4 0.090mg / L] for 1h, 2h, 4h and 8h of flooding treatment, and the depth of flooding should ...
Embodiment 2
[0035] Example 2: Gene structure and function prediction of zm-bRLZ
[0036] (1) Rapid amplification of cDNA ends (RACE) to isolate zm-bRLZ full-length cDNA
[0037] Trizol kit (purchased from Invitrogen, USA) was used to extract total RNA from the root system of Hz32 seedlings (for the operation method, refer to the manual of the kit) for the first strand synthesis of 5′-RACE and 3′-RACE cDNA, RACE technology The operation steps, reaction system and reaction program of the analysis were all in accordance with the operation method of the SMART RACE cDNA Kit amplification kit (purchased from Invitrogen, USA) referring to the kit’s manual. The primers were GSP1: TCCTAGTAAATCCAACCCTGTGAGC; NGSP1-1: TGCCTGTTTCTCTCCTCCTAGACCGCC; GSP2: TCCAGCAACTTTCAGATGCCAACCAA; NGSP2-1: GGGGTCCAACCGCAGGTACACGAGTA.
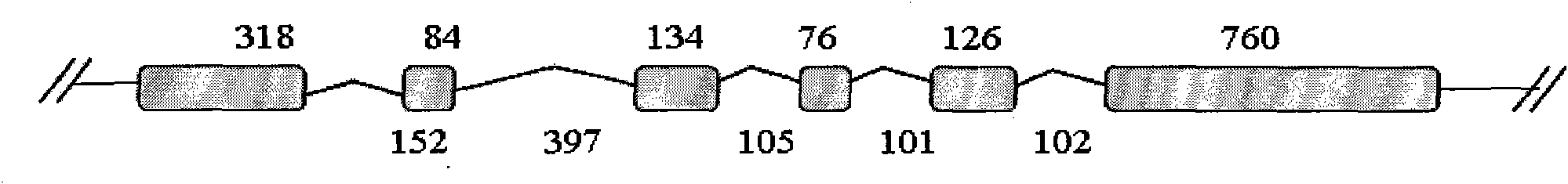
[0038] (2) Analysis of zm-bRLZ gene structure:
[0039] Based on the full-length cDNA, four pairs of primers (numbered bRLZ-1 to bRLZ-8, respectively, and the DNA sequences of the pr...
Embodiment 3
[0044] Example 3: In vitro expression of zm-bRLZ and analysis of its binding to anaerobic response elements
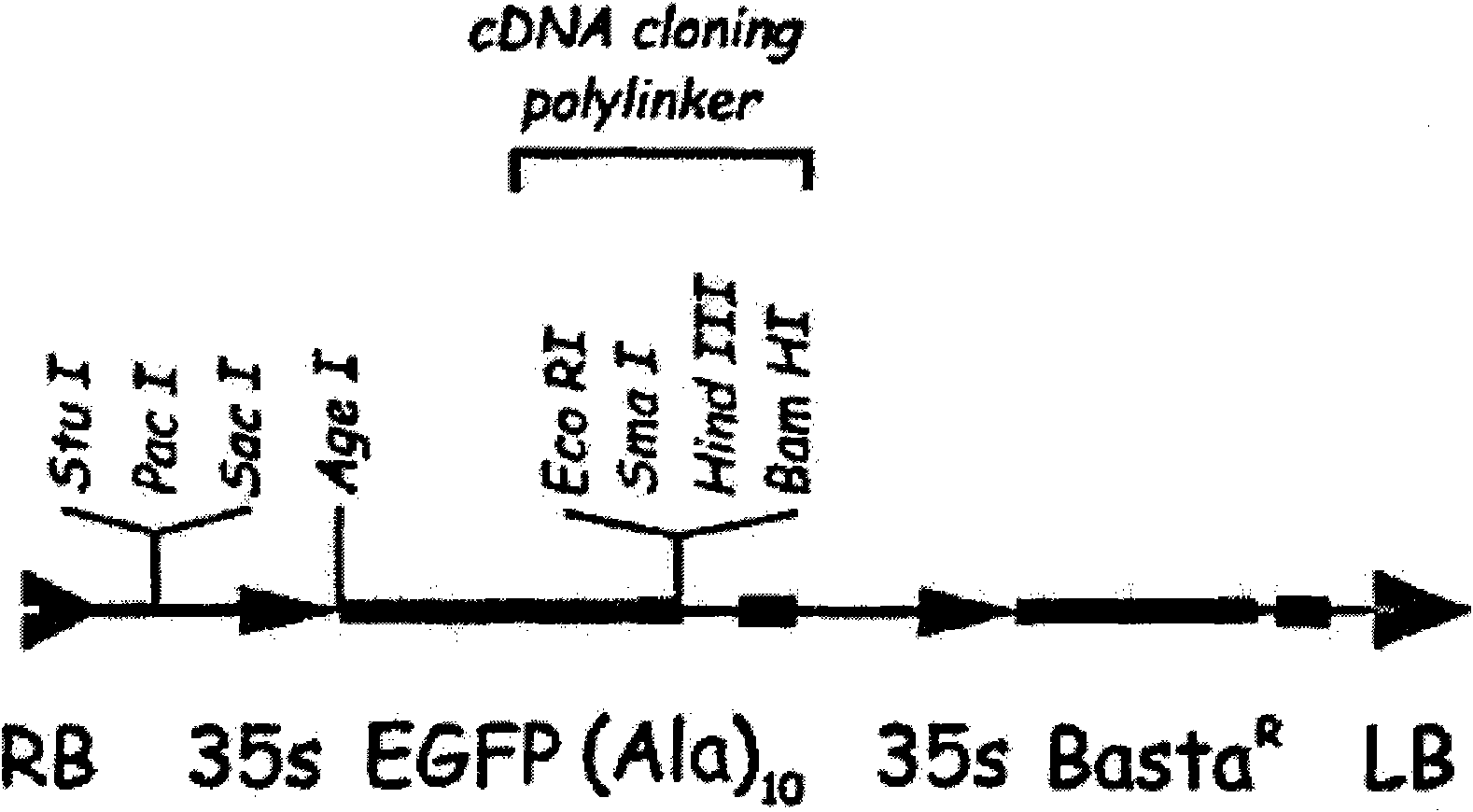
[0045] The upstream and downstream primers are: GTGGAATTCTAAGAAGTGCGCGTCGGAGCTG; TCAAGGCCAAATGTCCGCCGCCGAGCTCGA (where the underline is the restriction site), and the cDNA is used as a template for PCR amplification. The PCR product was recovered and purified, and inserted into the pGEX-KG vector ( Figure 5 ), construct the zm-bRLZ::GST structure, transform Rosseta host bacteria (Invitrogen, USA), and screen positive clones with ampicillin concentration of 100 μg / ml+LB plate. Add isopropylthiogalactopyranoside (IPTG) at a final concentration of 0.2mmoL / L to the positive clone LB liquid to induce at 4°C overnight, then centrifuge at 8000g for 10 minutes at 4°C to collect the precipitated bacteria. Take 1-2g bacteria and add 10mL breaking buffer (pH7.4, 50mM phosphate buffer: containing 0.5M NaCl, 0.5mg / mL lysozyme, 1mM phenylmethylsulfonyl fluoride PMSF, 1mM MgCl2, 1....
PUM
| Property | Measurement | Unit |
|---|---|---|
| Molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com