Gene contained in A.xylosoxidans LHB21 strain and used for coding catechol 2,3-dioxygenase
A catechol and dioxygenase technology, applied in genetic engineering, oxidoreductase, plant gene improvement, etc., can solve problems such as not suitable for cold regions, lack of cold resistance mechanism, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
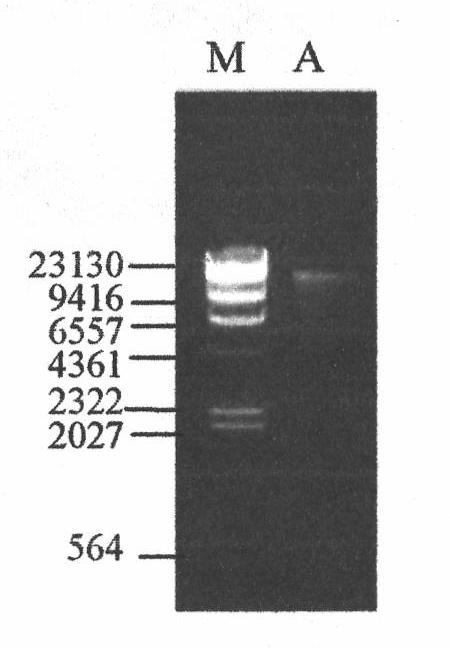
[0021] Example 1: Genomic DNA Extraction of Strain A.xylosoxidans LHB21
[0022] Use TaKaRa MiniBEST Bacterial Genomic DNA Extraction Kit Ver.2.0 (Code No.DV810A) to extract the genomic DNA of strain A.xylosoxidans LHB21, the specific operation is as follows:
[0023] 1. Bacterial culture: Pick a single colony and inoculate it into 1-4ml of LB (component concentration (W / V): peptone 1%, yeast extract 0.5%, NaCl 1%) liquid medium for overnight cultivation at 30°C.
[0024] 2. Take 1-4ml of supernatant, centrifuge at 10,000rpm for 2 minutes, and discard the supernatant.
[0025] 3. Fully suspend the bacterial pellet with 150ul of SP Buffer (containing RNase A1).
[0026] 4. Add 20ul of Lysozyme solution, mix well and let stand at room temperature for 5 minutes.
[0027] 5. Add 30ul of EDTA Buffer, mix well and let stand at room temperature for 5 minutes.
[0028] 6. Add 200ul of Solution A, shake vigorously and keep warm at 65°C for 10 minutes.
[0029] 7. Add 400ul of Solut...
Embodiment 2
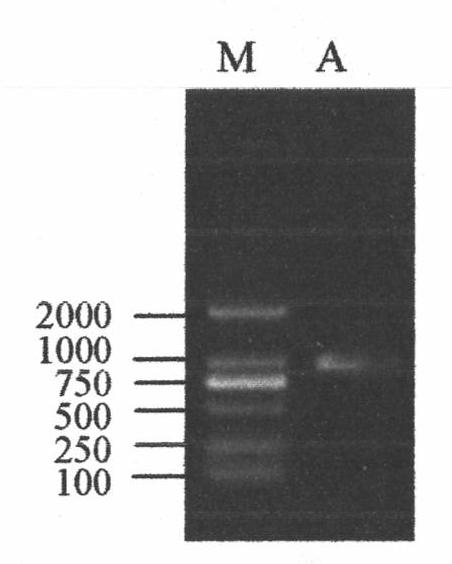
[0042] Embodiment 2: C23O gene PCR clone
[0043] 1. Cloning of C23O gene by PCR method
[0044] 1.1 Design and synthesis of primers
[0045] According to the C23O gene of A.xylosoxidans kf701 strain (as shown in SEQ ID NO: 3), the oligonucleotide primers used for C23O gene amplification were designed and synthesized, and the Nco I restriction enzyme was designed at the 5' end of the upstream primer F0 site, a Hind III restriction site was designed at the 5' end of the downstream primer R0.
[0046] PCR primers:
[0047] F 5'-ATGAACAAAGGTGTAATGCGAC-3' (SEQ ID NO: 4)
[0048] R 5'-TCAGGTCAGCACGGTCATGAA-3' (SEQ ID NO: 5)
[0049] YF2 5'-TGTTCACCAAGGTGCTCGG-3' (SEQ ID NO: 6)
[0050] YR2 5'-GGTCGAAGAAGTAGATGGTC-3' (SEQ ID NO: 7)
[0051] YF3 5'-GCCTCCATCATGTGTCCTTC-3' (SEQ ID NO: 8)
[0052] YR3 5'-TGTGTCGGTCATGGAGATCA-3' (SEQ ID NO: 9)
[0053] R02 5'-TCAGGTCAGCACGGTCATGAATCGTTCGTTGAGAAT-3' (SEQ ID NO: 10)
[0054] F0 5′- CCATGG CTATGAACAAAGGTGTAATGCGAC-3' (SEQ ID NO:...
Embodiment 3
[0079] Example 3: C23O gene expression
[0080] With pET-32a (+) (structure see Figure 4 ), construct the fusion gene expression vector pDSFL01 (see the structure Figure 5 ). Sequencing results showed that the C23O gene on the pDSFLO1 expression vector was a full-length C23O gene.
[0081] Pick a single colony of the positive expresser and inoculate it in 2ml liquid medium containing 100lg / ml ampicillin at 37°C, 200rpm and culture overnight. In 6ml LB medium, insert 100μl of the culture of Escherichia coli JM109 containing pDSFLO1 and grow to OD 600 0.55-0.7, add IPTG to make the final concentration reach 1.0mM, and induce expression at 30°C for 3 hours. Collect bacteria for electrophoresis analysis. SDS-PAGE electrophoresis results (see Image 6 ), Image 6 Middle M: Protein MW marker (Broad) (kDa), A: pET-32a(+) whole cell, B: pET-32a(+) supernatant, C: pET-32a(+) pellet, D: pDSFL01 whole cell , E: pDSFLO1 supernatant, F: pDSFLO1 pellet.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com