Cephalosporin acylase mutant and encoding gene and application thereof
A technology of cephalosporin acylase and sporin acylase, applied in application, genetic engineering, plant genetic improvement, etc., can solve the problem of low CPC activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1, the acquisition of cephalosporin acylase mutant
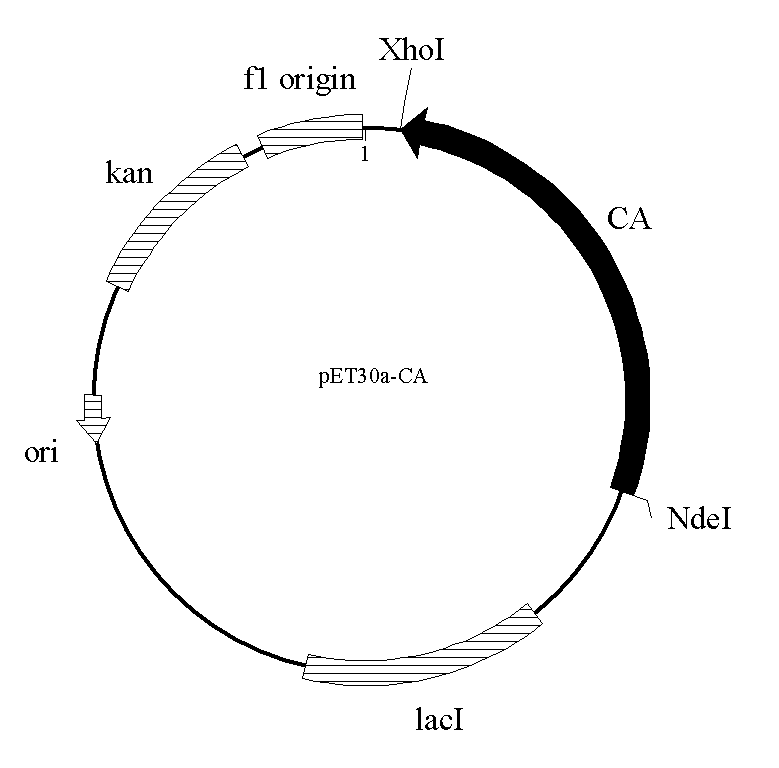
[0038] 1. Expression vector pET30(a)-CA-WT containing wild-type cephalosporin acylase CA
[0039] The amino acid sequence of the cephalosporin acylase of Pseudomonas SE83 acyII (GenBank: AAA25690.1, the amino acid sequence is sequence 1) was queried by NCBI (www.ncbi.nlm.nih.gov / ), and the method of overlapping PCR was used to synthesize Nucleotide sequence of cephalosporin acylase gene suitable for expression in E. coli. Specifically, its full-length amino acid sequence is input into the DNAworks program (http: / / helixweb.nih.gov / dnaworks / ) online. By setting the codon preference, DNAWorks outputs the nucleotide sequence fragments suitable for the cephalosporin acylase gene expressed in E.coli, each length is about 45bp, a total of 78 overlapping oligonucleotide sequences . The synthesized oligonucleotide fragments were dissolved with 10 mM Tris-HCl, pH 8.0 buffer. The PCR reaction system is in a 0.5ml ...
Embodiment 2
[0088] Example 2, Activity detection and conversion rate of cephalosporin acylase mutant
[0089] 1. Activity detection of cephalosporin acylase mutants
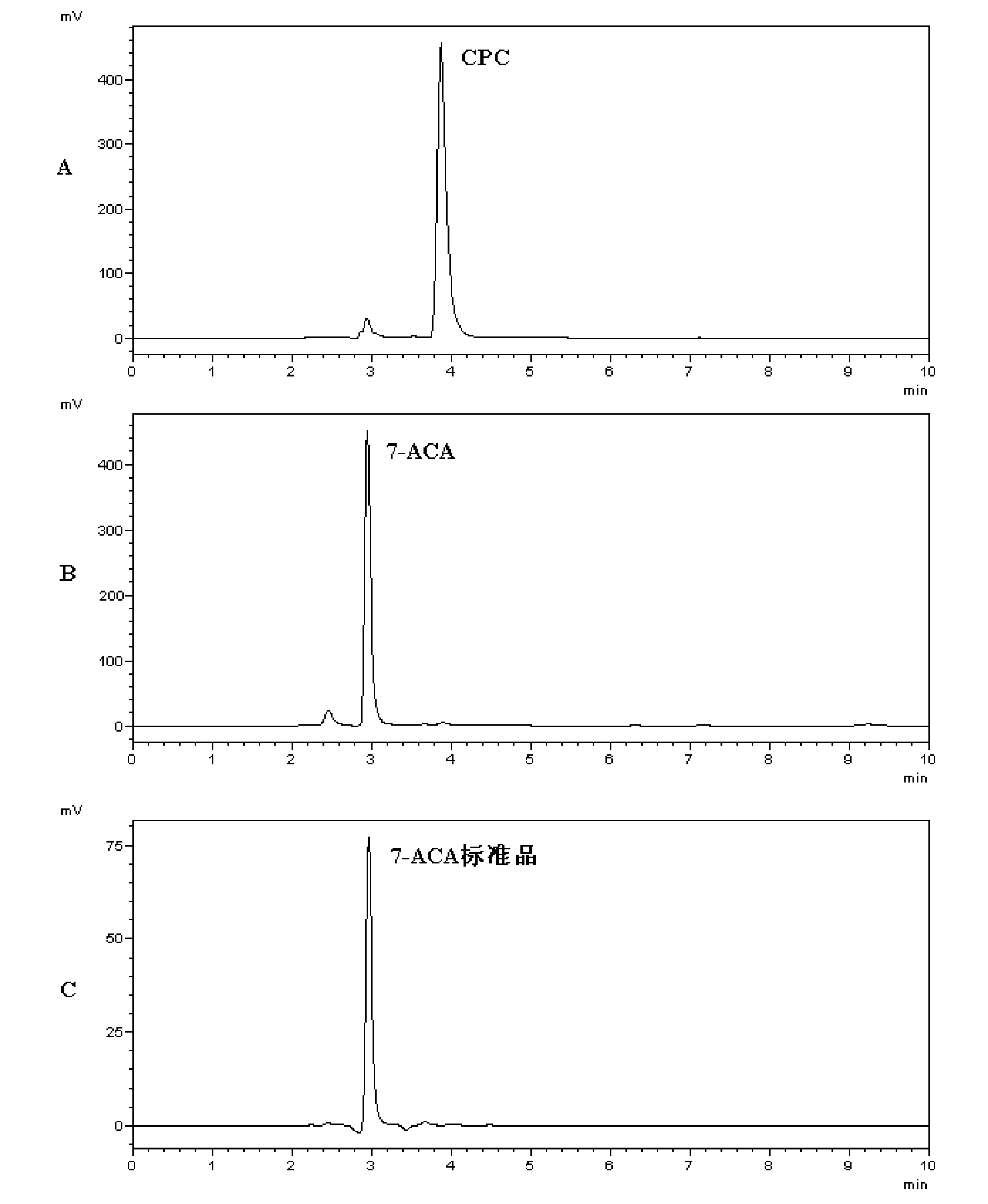
[0090] Take 20 μL of the purified CA-1C and CA-113 obtained in Example 1, respectively, and mix with pH=8.0, 100 mM Tris-HCl containing 3% (mass volume ratio) cephalosporin C (CSPC) 180 μl of buffer solution were mixed to obtain a pre-reaction mixture, and after reacting at 25° C. for 10 min, 200 μl of 40% glacial acetic acid was added to terminate the reaction to obtain a reaction product. Purified CA (wild-type cephalosporin acylase) was used as a positive control to obtain a positive control reaction product.
[0091] The pre-reaction mixture and products were detected by high-performance liquid chromatography, wherein Dima C18 liquid chromatography column 250mm×4.6mm was used, and the mobile phase consisted of 15% chromatographic methanol, 7.5% chromatographic acetonitrile, and 1% glacial acetic acid. 7-Aminocephalospo...
Embodiment 3
[0102] Embodiment 3, the mutant of cephalosporin acylase (CA) undergoes the construction of the derivative protein of amino acid residue substitution and activity determination
[0103] The idea is to start from the pET30(a)-CA-1C obtained in step 3 of the above-mentioned embodiment 1, and use the technique of overlapping PCR to introduce the Serβ471Ala mutation on the basis of CA-1C.
[0104] Using the pET30(a)-CA-1C obtained in Step 3 of Example 1 as a template, first use CA-For (the sequence is the same as in Example 1, Step 2) and CA-β471Lower: 5'-CATAACG AGC CAGCGCGCCATA-3' (where the underlined part is the mutation introduction site) was amplified by PCR at an annealing temperature of 60°C to amplify the upstream fragment of the mutant Serβ471Ala site;
[0105] Again, using pET30(a)-CA-1C as a template, use CA-Rev (the sequence is the same as in Example 1, step 2) and CA-β471Upper: 5'-GGCGCGCTG GCT CGTTATGT 3' (where the underlined part is the mutation introduction si...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com