PCR detection method of salmonella and primer pair
A Salmonella and detection method technology, applied in the detection field, can solve problems such as unfavorable diagnosis of etiology, time-consuming and laborious, complicated operation, etc., and achieve the effects of specific detection results, simple result judgment, and reduction of detection costs.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1. Select the conserved sequence in the Salmonella genomic DNA sequence and design primers
[0028] The base sequence selected from the genome DNA sequence of Salmonella, such as the sequence shown in SEQ ID NO: 1, is a conserved sequence. Then use the Primer Premier 5.0 software to design primers, and the primer sequences are as follows:
[0029] The base sequence of the forward primer is: gagttatga accaacagct (SEQ ID NO: 2);
[0030] The base sequence of the reverse primer is: gccgctaaac tacacgatga (SEQ ID NO: 3);
[0031] Primers were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0032] 2. Extract the DNA of the sample to be tested and amplify it by PCR;
[0033] Refer to the "People's Republic of China Import and Export Commodity Inspection Industry Standard Export Food Salmonella" (SN 0170-92) for enrichment culture of oysters to be inspected.
[0034] Inoculate each bacterial culture into 30mL lysing broth (LB) liquid medium, sha...
Embodiment 2
[0038] Embodiment 2, broad spectrum and specificity test
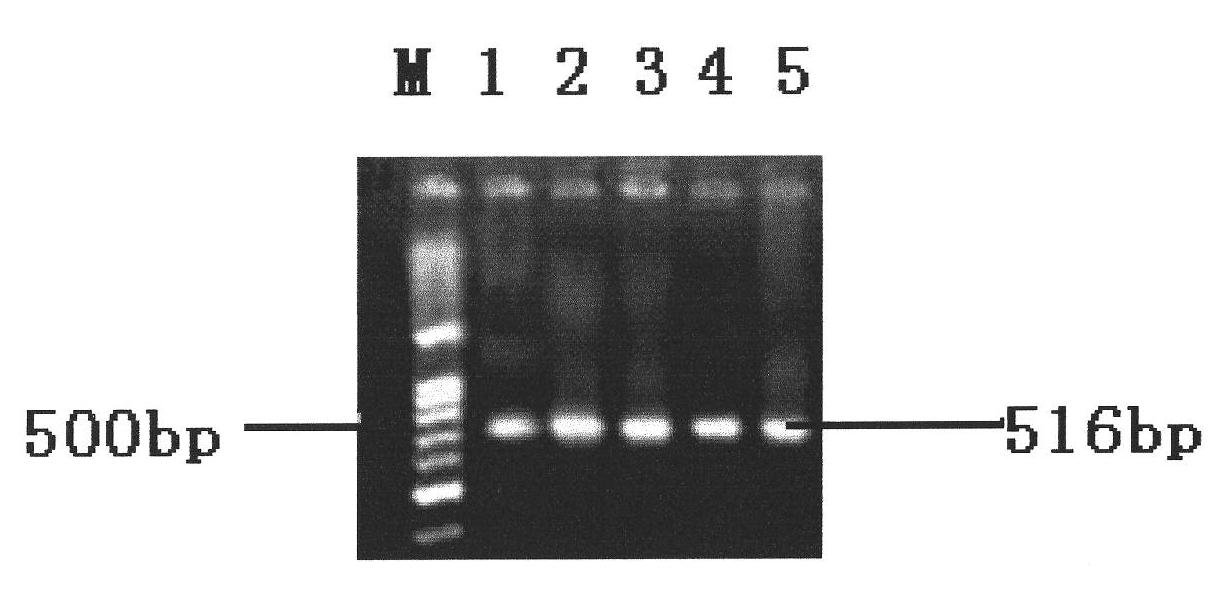
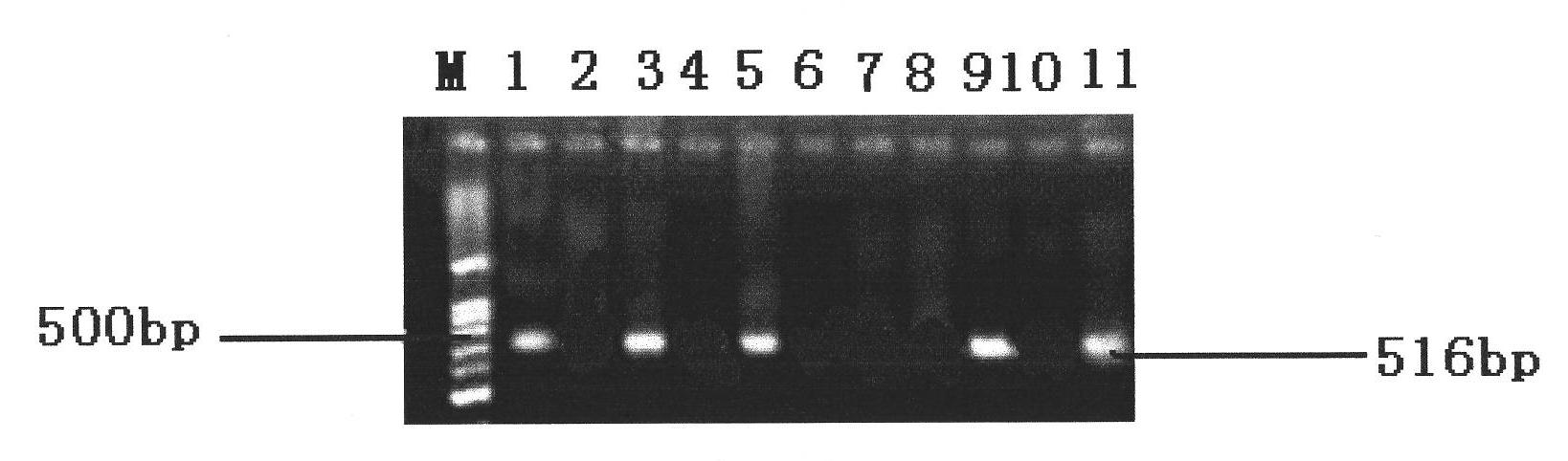
[0039] Different serotypes of Salmonella and other non-Salmonella standard strains were purely cultured, and the DNA template was extracted according to the method in Example 1, PCR amplification was performed, and the amplified products were detected by electrophoresis. see results figure 2 and image 3 , figure 2 The strains represented by each lane are: 1. S.indiana; 2. S.enterilids; 3. S.derby; 4. S.typhimurium; 5. S.senftenberg; 6. S.kentucky; 7. S.cholerae suis ;8, S.hartford; image 3 The strains represented by each lane are: 1. S.hartford; 2. E.coli; 3. S.Indiana; 4. Shigella; 5. S.cholerae suis; 6. Staphyloccocus; 7. S.typhimurium; 8. Citrobacter 9. S. enteritids; 10. Enterobacter cloacae; 11. S. derby. The results showed that the 8 different serotypes of Salmonella could amplify the bands, but none of the non-Salmonella could amplify the bands. It can be seen that the detection method in Example 1 has ...
Embodiment 3
[0040] Embodiment 3, sensitivity test
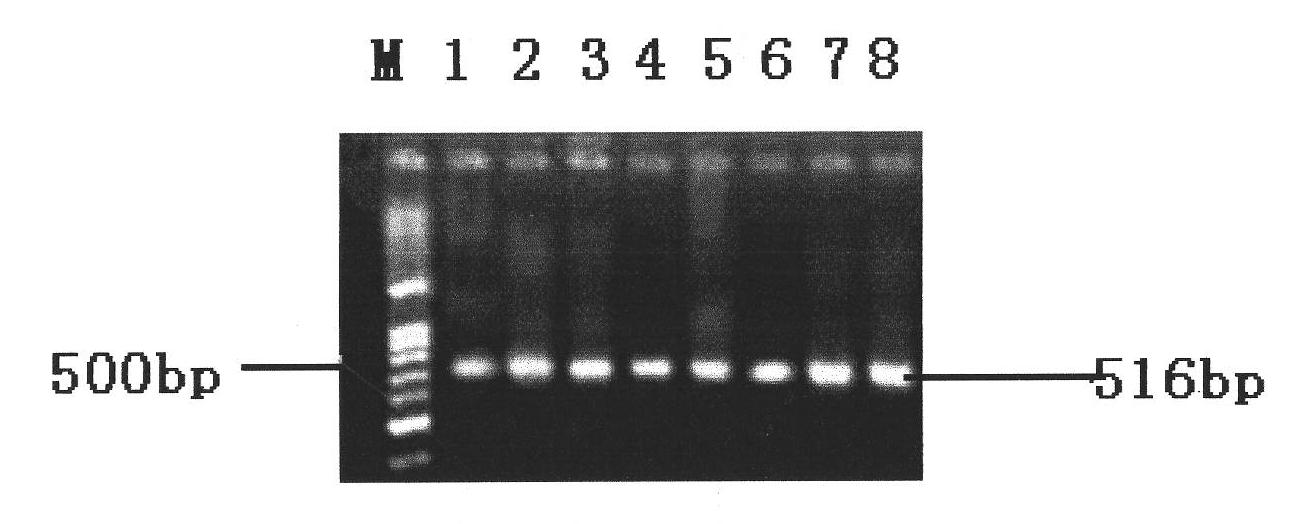
[0041] Take 1 mL of Salmonella culture to extract the DNA template and measure the concentration, and then dilute it to different concentrations in a 10-fold gradient. Take 1 μL of each concentration for PCR amplification, and detect the amplified product. The detection method refers to Example 1. The liquid culture of Salmonella was diluted 10 times, and the DNA template was extracted for amplification detection, and a plate colony counting was performed at the same time. The result is as Figure 4 as shown, Figure 4 Middle, 1, undiluted template; 2, 10 times diluted; 3, 10 2 Double dilution; 4, 10 3 Double dilution; 5, 10 4 1-fold dilution; 6, 10 5 double dilution. It can be seen from the figure that diluting the concentration of the extracted DNA template to 10 4 When multiple times, the target DNA band can still be clearly observed. The DNA template was detected by ultraviolet light, and the concentration of the stock soluti...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com