Kit for jointly detecting four deafness predisposing genes and application thereof
A joint detection technology of susceptibility genes for deafness, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, etc., can solve the problems of many false negatives and false positives, difficult interpretation of results, cumbersome operation, etc., to prevent false positives and false negative, increase Tm value, shorten the effect of primer length
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
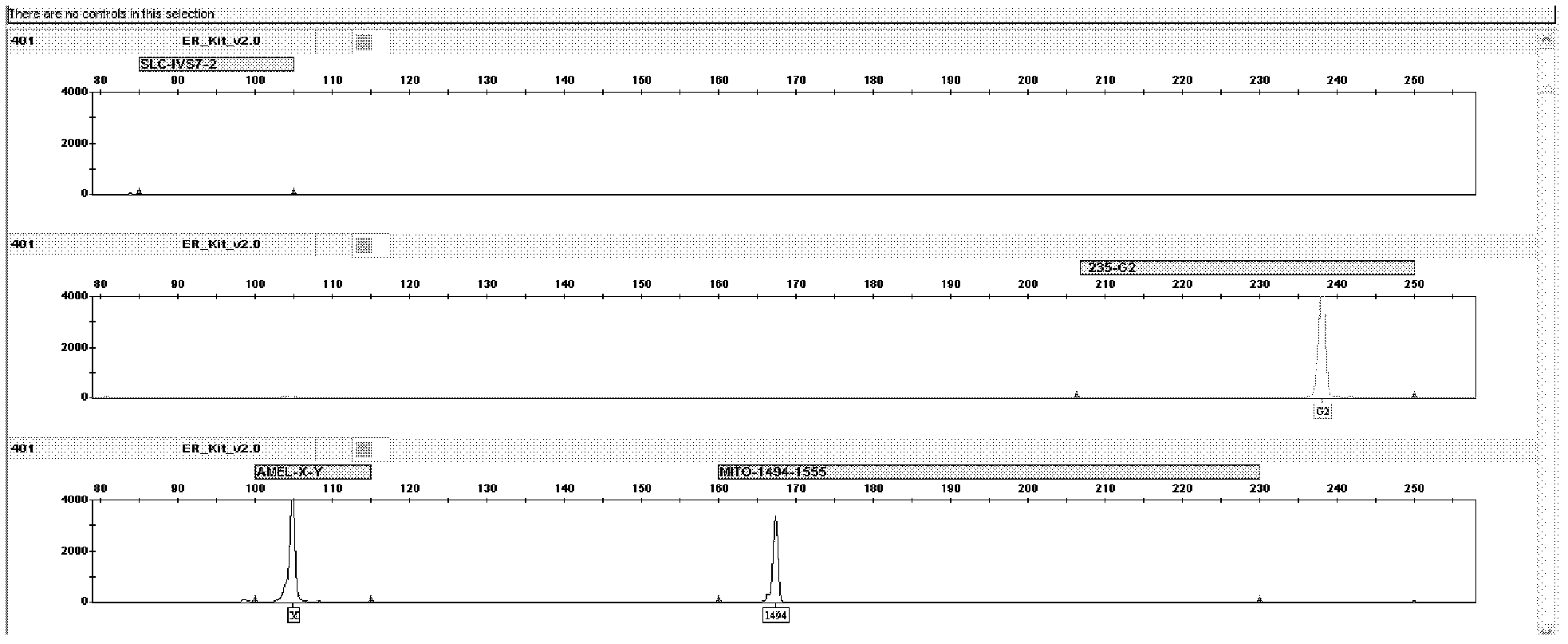
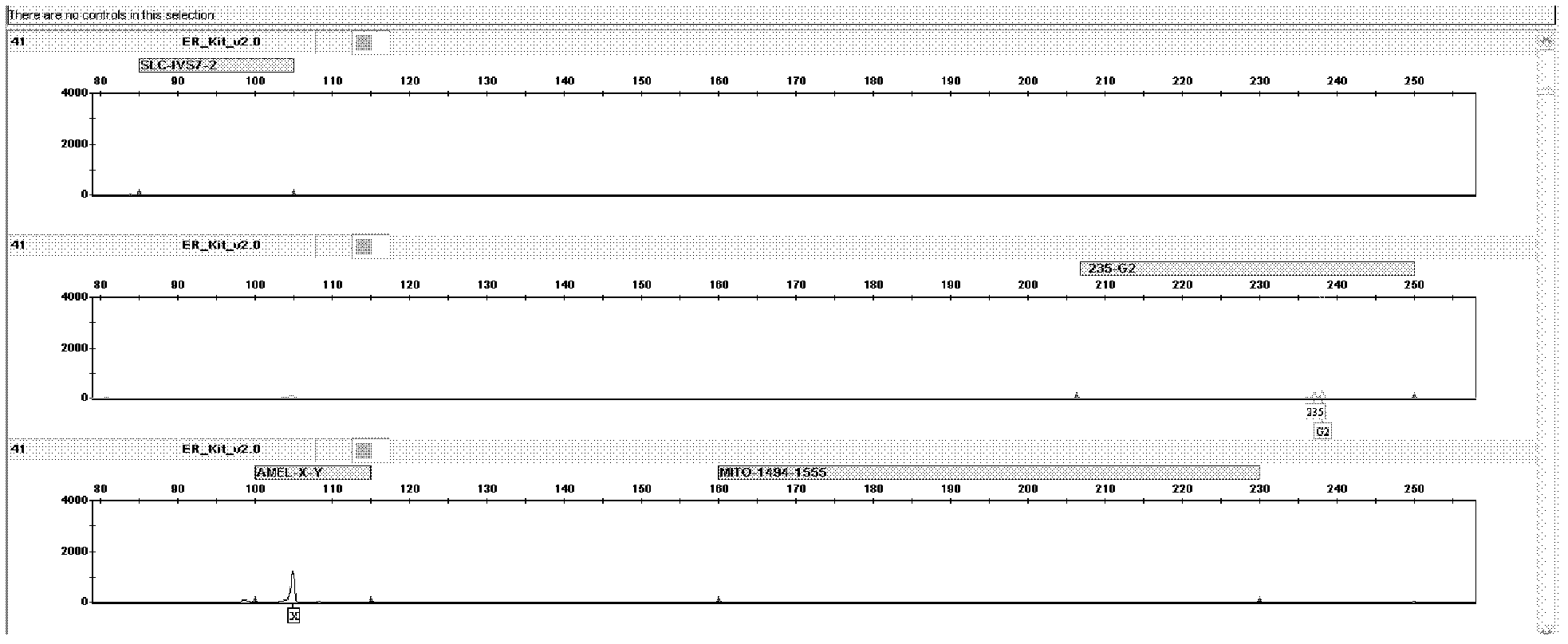
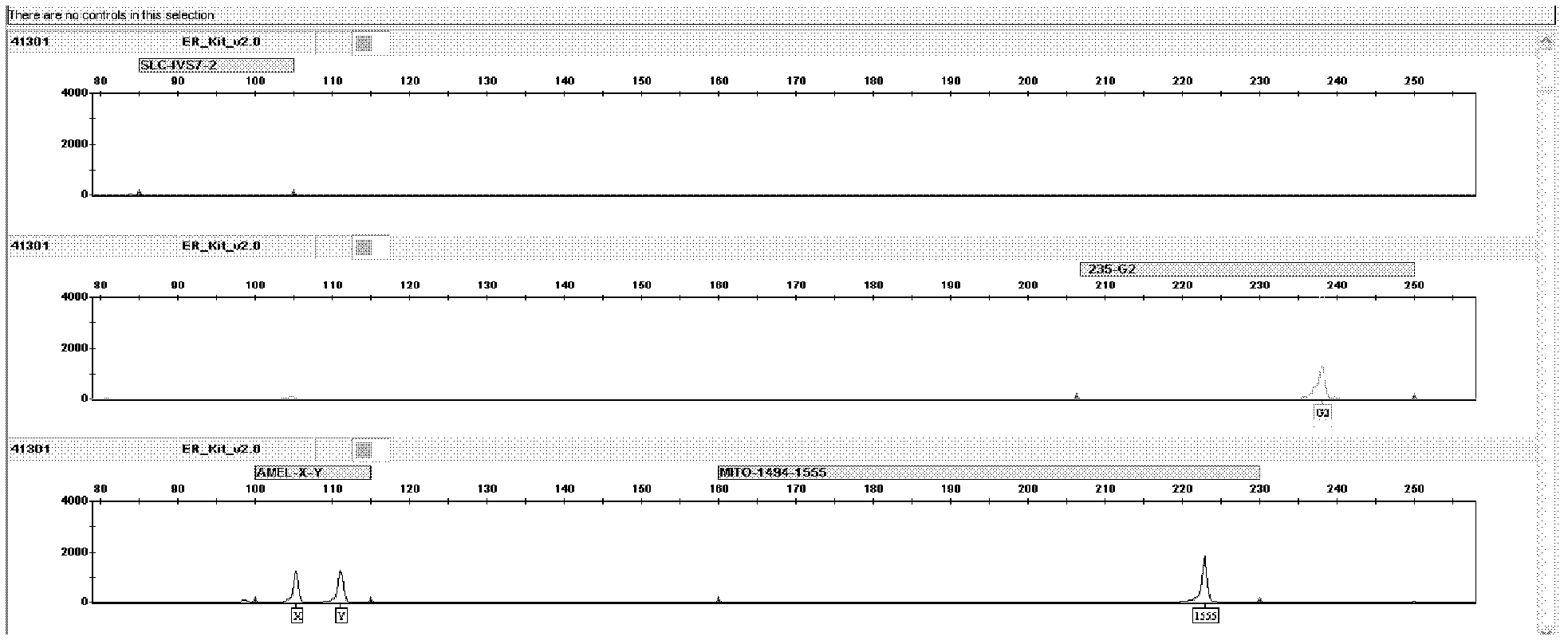
[0044] The kit of the present invention detects mutations and DNA samples of normal individuals. The primers for the detection of the deaf susceptibility gene SLC26A4 mutation hotspot IVS7-2A>G are labeled with blue fluorescent dye, and the primers for GJB2235delC detection are labeled with green fluorescent dye, and are used for the Amelogenin locus, 12S rRNA 1494C>T, 1555A>G The primers for mutation detection are labeled with yellow fluorescent dye, and the internal standard is labeled with red fluorescent dye.
[0045] 1. The 1,000 samples to be tested have all been sequenced and detected by using the technical method of "DNA extraction-PCR amplification-sequencing". Among them, there were 10 samples of IVS7-2A>G mutation, 10 samples of GJB2235delC mutation, 10 samples of 12SrRNA 1555A>G mutation, and 1 sample of 1494C>T mutation.
[0046] 2. Genomic DNA extraction from samples
[0047] Chelex extraction method: Cut off 1-3mm blood spot and put it in a 1.5mL centrifuge tu...
Embodiment 2
[0065] The unmodified primers were used to detect DNA samples from normal individual blood spots at the same concentration (5.0ng / 25uL system). The labeled primers are the same as in Example 1. The non-labeled primers used for the detection of deaf disease susceptibility loci were the common primers corresponding to the primers in Example 1 without LNA modification.
[0066] 1. The sample to be tested is the same as the normal individual blood spot in Example 1.
[0067] 2. Genomic DNA extraction from samples
[0068] Genomic DNA was extracted by Chelex method.
[0069] 3. Detection and analysis of amplification and amplification products
[0070] 3.1PCR amplification system:
[0071]
[0072] 3.2 PCR amplification procedure: Same as Example 1.
[0073] 4. Fluorescent detection of the amplified product on a genetic analyzer
[0074] With embodiment 1.
[0075] 5 Conclusion
[0076] The result is as Figure 6 As shown, for male normal individuals, the G2 position (2...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com