Quick identification method and application of transgenic fish homozygote
An identification method and a transgenic technology, applied in the field of aquatic animal genetics and breeding, can solve the problems of time-consuming and laborious, low stability of identification results, and achieve the effects of speeding up the breeding process, excellent breeding traits, and stable genetic traits.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] A method for identifying homozygous transgenic fish, the steps of which are:
[0031] 1. DNA extraction
[0032] Cut about 0.5-1cm 2 Genomic DNA of transgenic fish was extracted from caudal fin tissue of GH transgenic fish according to the operation guide of "Blood / Cell / Tissue Genomic DNA Extraction Kit" (product of Beijing Tiangen Biotechnology Company).
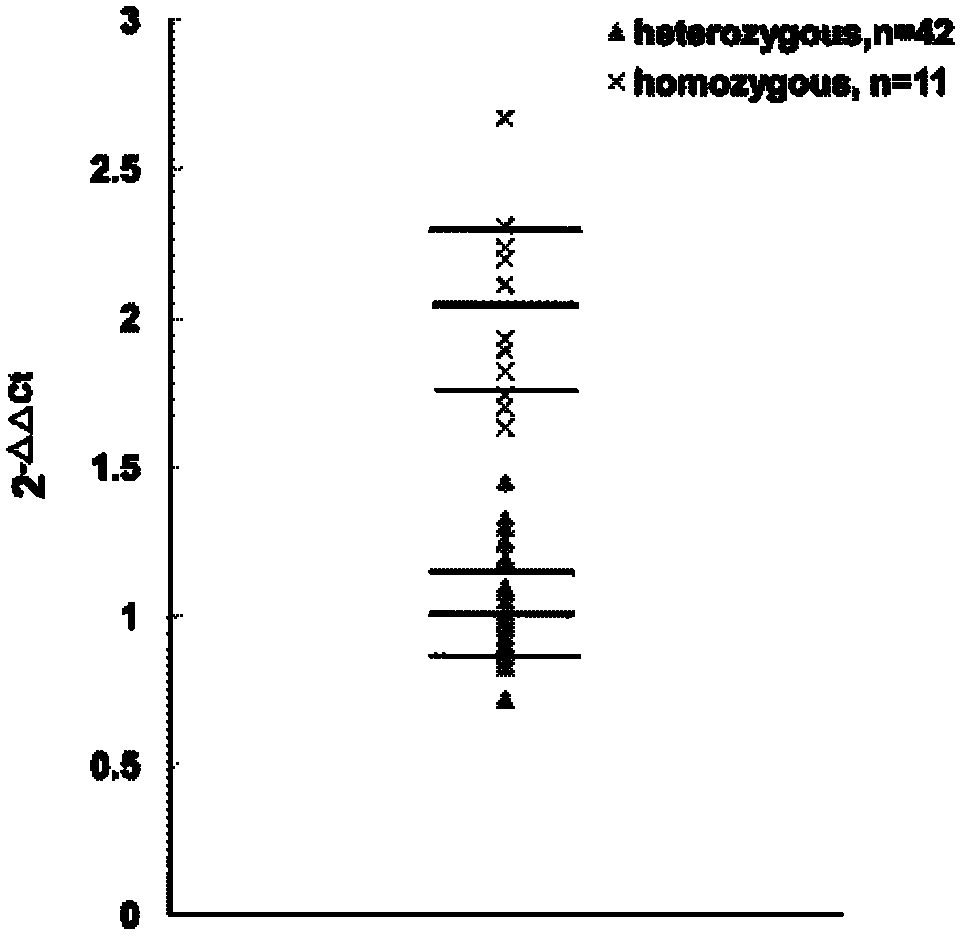
[0033] 2. Positive identification of transgenic fish
[0034] Positive identification of GH transgenic fish was carried out by conventional PCR technique. The total volume of the PCR system is 25ul, including 20ng DNA, 100uMdNTP, 2.5ulBuffer (25mM), 2.5ul MgCl 2 (25 mM), 1 Unit T. Aquaticus DNA polymerase (Fermentas, USA), and 1 μl each of primers (10 mM) (Invitrogen, the same below). Primer sequences: GHf (5'-CATTT ACAGTTCAGCCATGGCTAGA-3') and GHr (5'-AGCACCACCGACAACAGCACTA ATG-3'). Reaction program: pre-denaturation at 94°C for 5 minutes, amplification for 30 cycles (94°C, 30s; 58°C, 30s; 72°C, 30s), and final...
Embodiment 2
[0042] Application of a screened homozygous transgenic Yellow River carp in cultivating excellent cultured fish species. The specific process is:
[0043] A, the identified male transgenic Yellow River carp homozygote and the female transgenic Yellow River carp homozygote are cultured in ponds according to conventional methods, and after they develop to sexual maturity, artificial induction of labor is carried out according to conventional methods;
[0044] B. artificial insemination of the homozygous semen of the male transgenic Yellow River carp and the eggs of the female transgenic Yellow River carp;
[0045] C. The progeny obtained by self-crossing is a homozygous family of transgenic fish with stable genetic properties, and the obtained homozygous family of transgenic Yellow River carp is bred and cultivated in ponds according to conventional methods, thereby cultivating new varieties of excellent cultured fish.
[0046]
[0047]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com