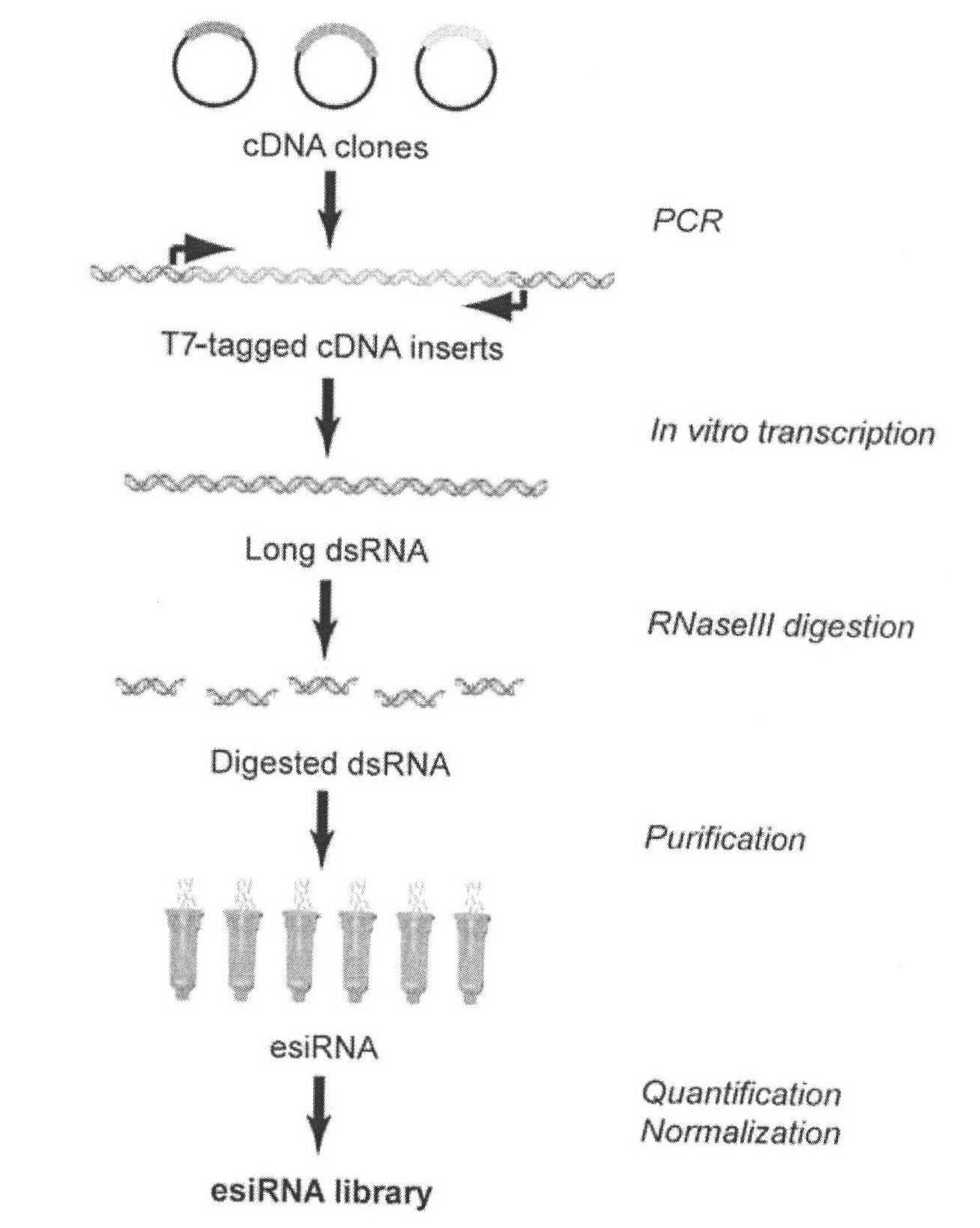
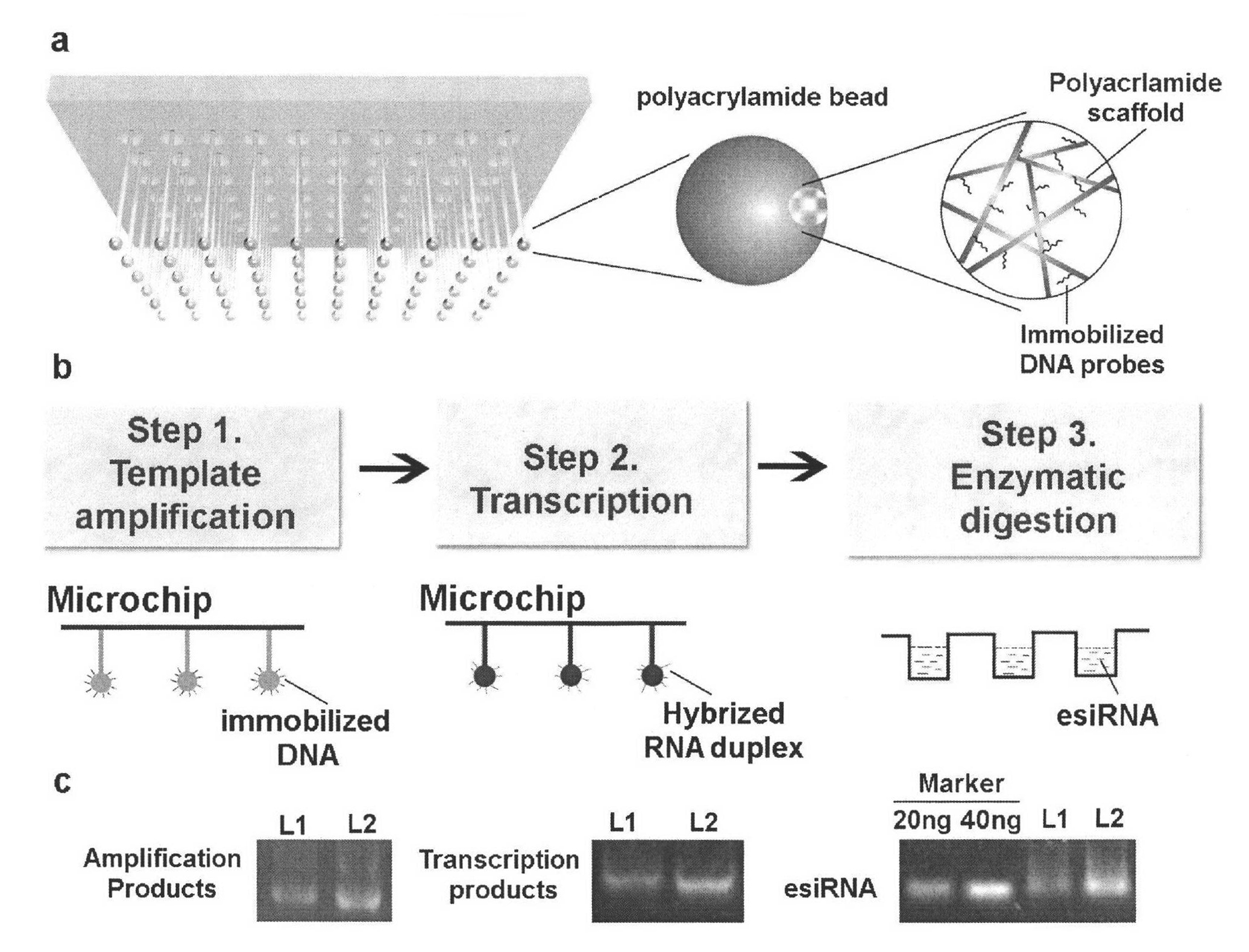
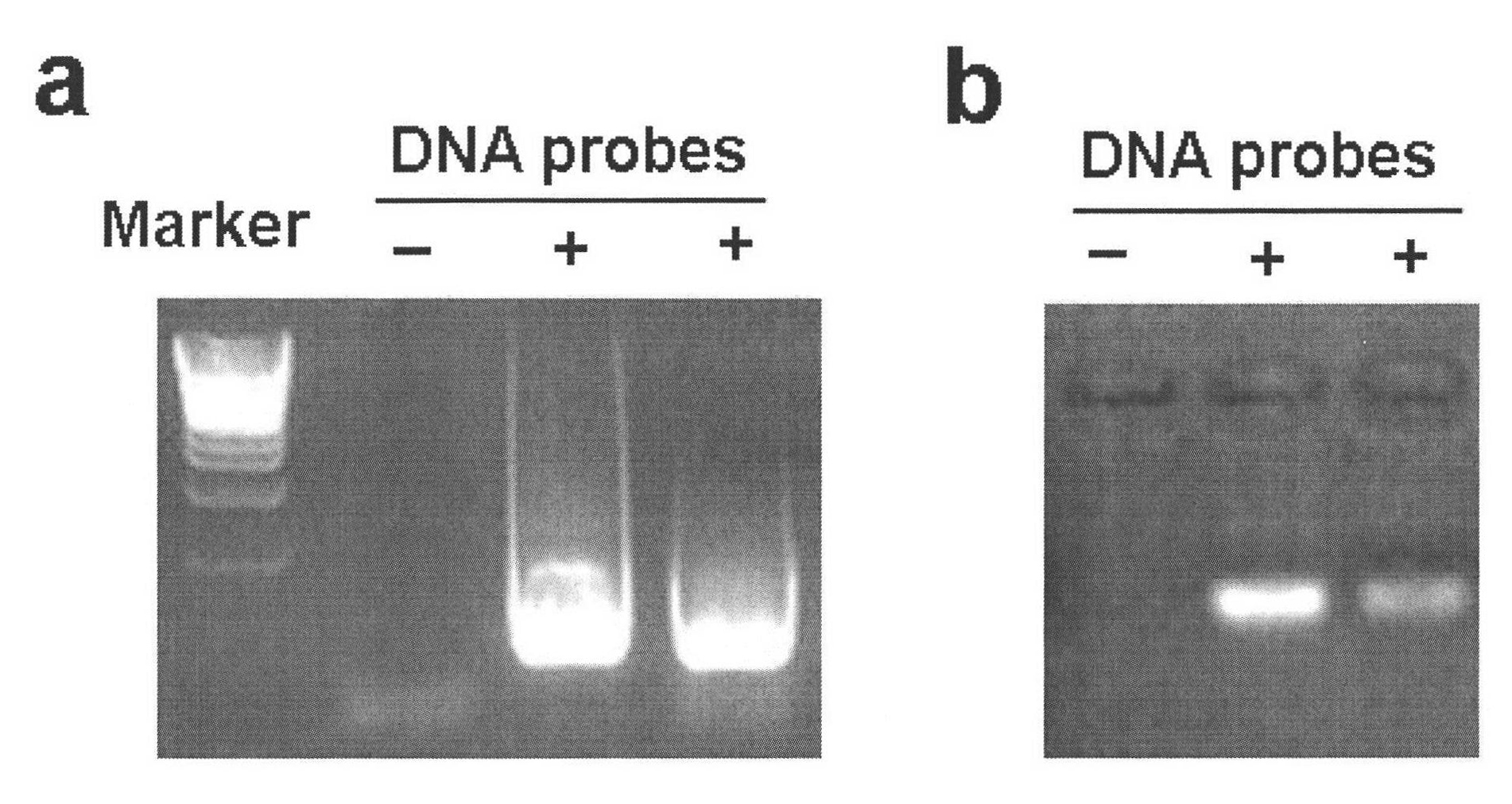
Method for simply and quickly preparing ready-to-use esiRNA
A DNA probe and polymer block technology, applied in the field of small RNA preparation, can solve problems affecting the preparation and application of esiRNA
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0032] 1. Preparation of polymer blocks:
[0033] Acrylamide (acrylamide:bisacrylamide=19:1) 10wt% glycerol solution, 0.1M sodium phosphate buffer solution (pH=7.0), 3wt% methacrylic acid, 0.005wt% APS and 2mM TEMED was mixed well under a nitrogen atmosphere, then transferred to test tubes and incubated overnight at 65°C to allow polymerization.
[0034] 2. Immobilization of DNA probes on polymer blocks:
[0035] Use MES (0.1M) buffer solution (pH=6.0) containing EDC (2mM) and sulfo-NHS (2mM) to activate the carboxyl group on the polyacrylamide polymer block, and then add the 5'-terminal amino-modified DNA Oligomer (Invitrogen Co. Ltd., Shanghai) and incubated overnight at room temperature. The sequence of the DNA probe used in the PCR amplification reaction (that is, the probe immobilized on the first nucleic acid chip) is as follows: 5'-amino-TCACAGGATGGCTAATACGACTCACTATAGGGC-3', and the DNA probe used in the transcription process (that is, the immobilized The sequence of...
Embodiment 1
[0051] The bottom of the 96-well plate commonly used in the laboratory was pierced to form small holes, and a capillary was inserted into each small hole and fixed with PDMS, and polyacrylamide microspheres of the same volume and size were fixed at the lower end of the capillary. The first nucleic acid chip and the second nucleic acid chip can be obtained by immobilizing different DNA probes on polyacrylamide, wherein the DNA probe sequence on the first nucleic acid chip can be complementary to a sequence of the PCR amplification product DNA molecule, and the second The DNA probe sequence on the two-nucleic acid chip can be complementary to a sequence of transcription product RNA. Immerse the first nucleic acid chip into the PCR amplification solution to carry out the PCR amplification reaction. After one PCR reaction, take out the chip and rinse it, and then add another set of PCR amplification solutions that do not contain DNA templates to carry out the PCR amplification reac...
Embodiment 2
[0055] The experimental process is the same as in Example 1, except that the materials for preparing polymer microspheres are changed to polymethacrylamide, polylactic acid (PLA), polylactic acid-glycolic acid polymer (PLGA), polyacrylic acid (PAA), polyformaldehyde Acrylic acid, poly 2-hydroxyethyl (meth) acrylate, poly N-isopropyl (meth) acrylamide, polyvinyl acetate or polyacrylamine, the results obtained are similar to those of Example 1.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com