Construction method for prokaryotic secretory expression vector of protein transduction domain-Apoptin (PTD-Apoptin) fusion protein and application of prokaryotic secretory expression vector
A fusion protein, secretory expression technology, applied in the field of microorganisms, can solve the problems of restricted activity, protein does not meet the requirements, and the renaturation efficiency of inclusion bodies is low, and achieves the effect of optimizing expression conditions.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
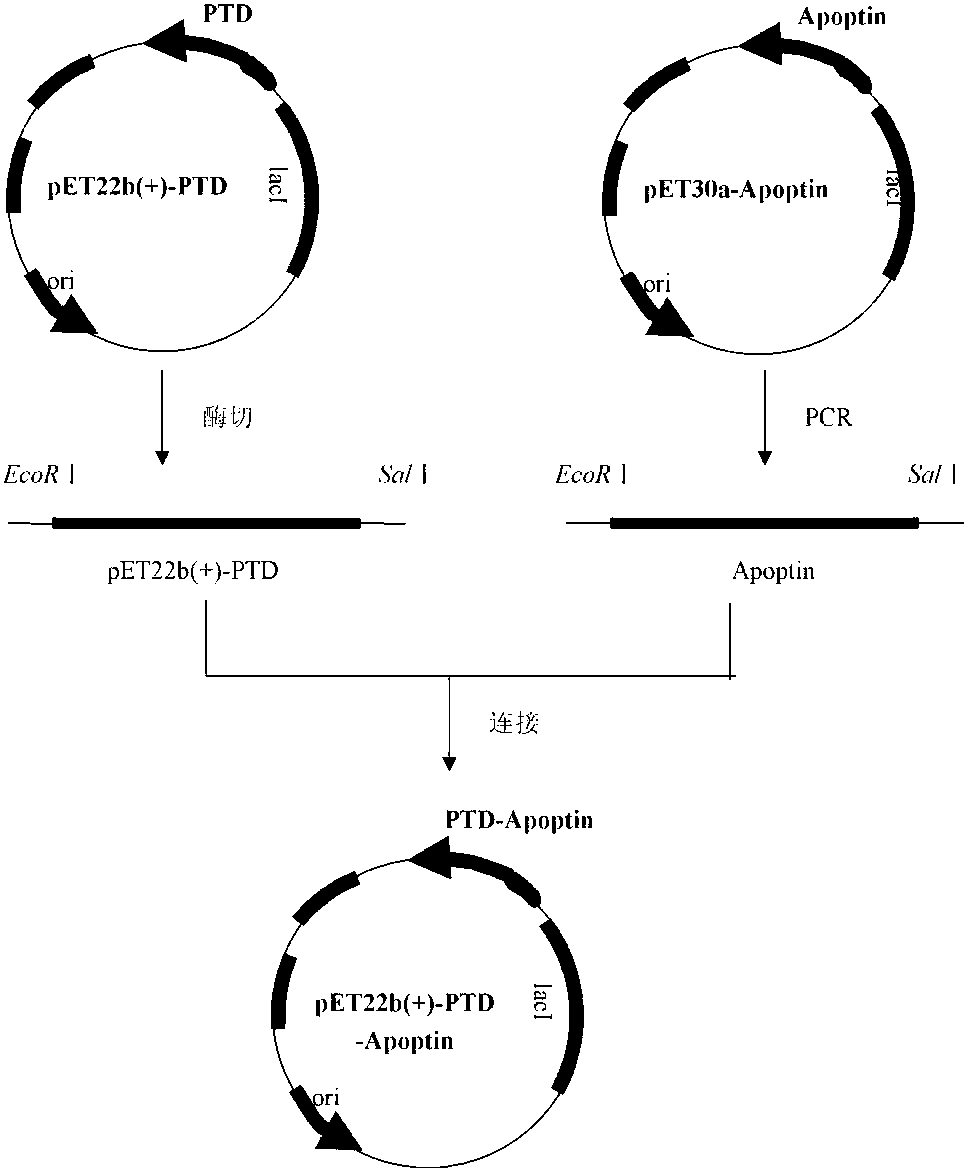
[0032] Example 1 Construction and secretory expression method of PTD-Apoptin fusion protein
[0033] (1) Materials and methods
[0034] 1. Material: T 4 DNA ligase, restriction enzyme Bam H I. EcoR I and Sal Ⅰ. Plasmid small extraction kits and gel recovery kits are all products of TaKaRa; IPTG (isopropyl-β-D-thiogalactopyranoside), MTT (thiazolium blue), DMSO (dimethyl sulfoxide) was purchased from SIGMA.
[0035] 2. Method
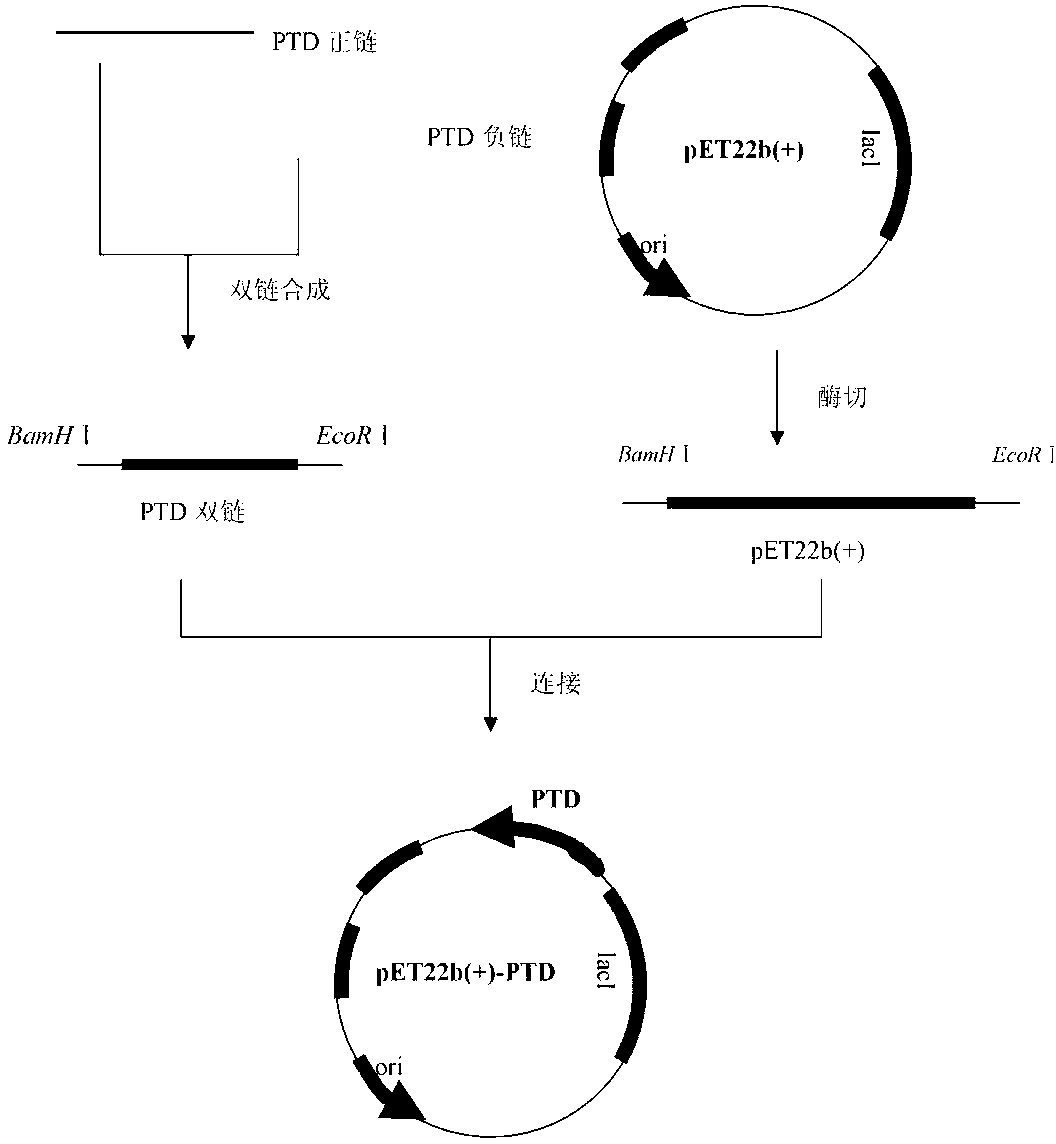
[0036] 2.1 PTD sequence preparation
[0037] The PTD sequence was artificially synthesized according to the method of primer design, and an enzyme cutting site was introduced at the 5' end of the positive strand of the PTD sequence Bam H Part of the recognition site of I, introduced at the 3' end EcoR Partial recognition site of I; introduction of a restriction site at the 5' end of the negative strand of the PTD sequence EcoR Part of the recognition site of I, introduced at the 3' end Bam H Part of the recognition site of I. After ann...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com