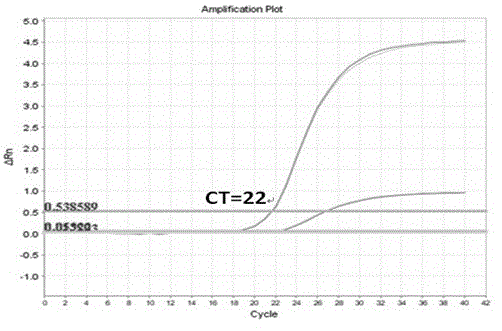
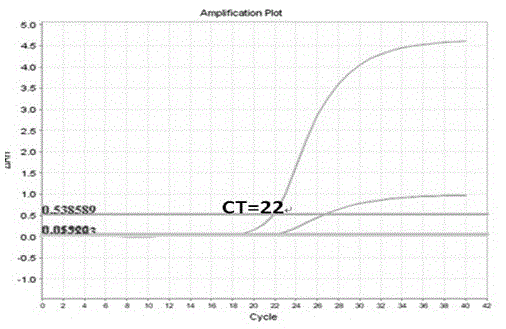
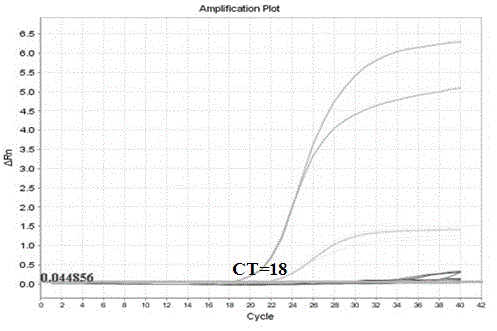
Human K-ras (K-rat sarcoma) gene mutation parting type fluorescent quantitation PCR (polymerase chain reaction) detecting reagent kit and detecting method
A detection kit and fluorescence quantitative technology, applied in fluorescence/phosphorescence, material excitation analysis, DNA/RNA fragments, etc., can solve the problems of operator hazard, high detection cost, complicated operation process, etc., to improve detection sensitivity, Get results with low quality requirements and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0077] Example 1 Detection of six mutations in codon 12 of the K-ras gene by fluorescent quantitative PCR (codons 12GGT>GAT, 12GGT>GCT, 12GGT>GTT, 12GGT>AGT, 12GGT>CGT, 12GGT>TGT)
[0078] Design specific ARMS forward primers that can specifically recognize and amplify the above 6 mutations, 6 ARMS forward primers share one downstream primer, Taqman-MGB detection probe, internal control primer pair and detection probe, each primer, The probe sequences are as follows:
[0079] The peptide nucleic acid (PNA) sequence is shown as SEQ ID NO: 1 in the sequence listing.
[0080] SEQ ID NO 1: H2N-CTACGCCACCAGCTC-CON2H.
[0081] For the 12GGT>GAT codon mutation, the forward ARMS primer sequence was designed as 5'-AACTTGTGGTAGTTGGAGCTGA-3', as shown in SEQ ID NO:2.
[0082] For the 12GGT>GCT codon mutation, the forward ARMS primer sequence was designed as SEQ ID NO3: 5'-ACTTGTGGTAGTTGGAGCTGC-3', as shown in SEQ ID NO:3.
[0083] For the 12GGT>GTT codon mutation, the forward ARMS...
Embodiment 2
[0094] Example 2: Detection of a mutation in codon 13 of the K-ras gene (codon 13GGC>GAC) by fluorescent quantitative PCR.
[0095] Design specific ARMS forward primers that can specifically recognize and amplify one of the above mutations, one ARMS forward primer shares one downstream primer, Taqman-MGB detection probe, internal control primer pair and detection probe, each primer, The probe sequences are as follows:
[0096] PNA sequence is H 2 N-CTACGCCACCAGTC-CON 2 H, as shown in SEQ ID NO1: in the sequence listing: SEQ ID NO1: H2N-CTACGCCACCAGCTC-CON2H.
[0097] For GGT>GAC 13 codon mutation, the designed primer sequence is SEQ ID NO8: 5'-TTGTGGTAGTTGGAGCTGGTGA-3', as shown in SEQ ID NO: 8. One ARMS forward primer sequence shares one reverse primer sequence SEQ ID NO9 5'-CAAGATTTACCTCTATTGTT-3', as shown in SEQ ID NO: 9 in the Sequence Listing. ;
[0098] The mutation detection probe is: SEQ ID NO10: 5'-FAM-GAGTGCCTTGACGAT-MGB-3' as shown in SEQ ID NO10 in the sequen...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com