Anti-HPV (human papillomavirus) medicine virtual screening method by using DNA helicase E1 of HPV as target point
A DNA helicase and human papillomavirus technology, applied in the field of virtual screening of new anti-HPV virus drugs, can solve problems such as low efficiency, a lot of manpower, time and energy, complex natural medicines and food components, etc. cycle, effect of increasing speed and efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Example 1 Using the tyrosine residue Y492 of HPV18 E1 protein as the active site, the virtual screening of anti-cervical cancer drugs was carried out.
[0036] (1) Virtual screening steps are as follows:
[0037] 1. Acquisition of the three-dimensional crystal structure of HPV18 E1 protein in Protein Data Bank
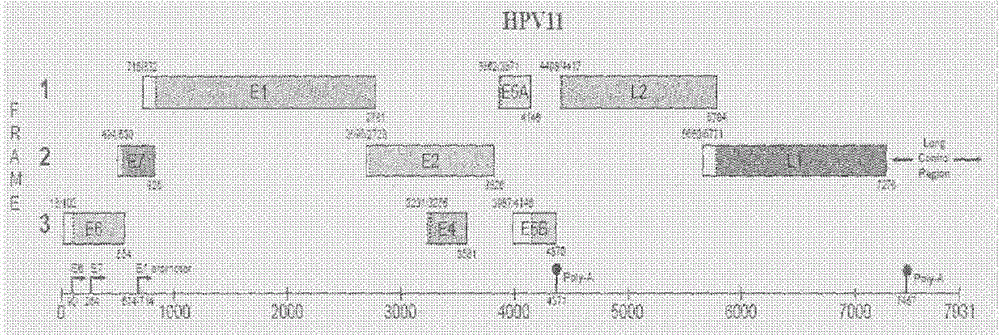
[0038] Using the Protein Data Bank (PDB) database, the three-dimensional crystal structure of the HPV18 E1 protein required for our experiment ( image 3 ) to download and get the amino acid sequence of E1 protein (FASTA format, figure 2 ), so as to carry out the next operation.
[0039] 2. Use Autodock molecular docking software to determine the active center and set the active pocket according to the active tyrosine residue site Y492 of E1
[0040] In this step, firstly, a known molecule that can undergo an addition reaction with the active tyrosine residue site Y492 of E1 is used as a positive control molecule. The binding energy of the interaction and t...
Embodiment 2
[0076] Example 2 A virtual screening of anti-HPV virus drugs by using R589 of the E1 protein as the active site to construct an active pocket.
[0077] The method is the same as in Example 1, the difference is: when setting the active pocket: the central coordinate of R589 is the center of the active pocket, and finally the conditions for setting the active pocket are as follows: the active tyrosine residue site R589 of E1 is used as the active pocket. Active pocket in the center: the center coordinates are 36.410 for x, 25.423 for y, and 106.056 for z, the grid size is 40×40×40, and the spacing is 0.375nm.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com