Glycerol-3-phosphate acyl transferase
An amino acid and polynucleotide technology, which is applied in the field of glycerol-3-phosphate acyltransferase, can solve the problems of inability to obtain supply and achieve the effect of high production efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
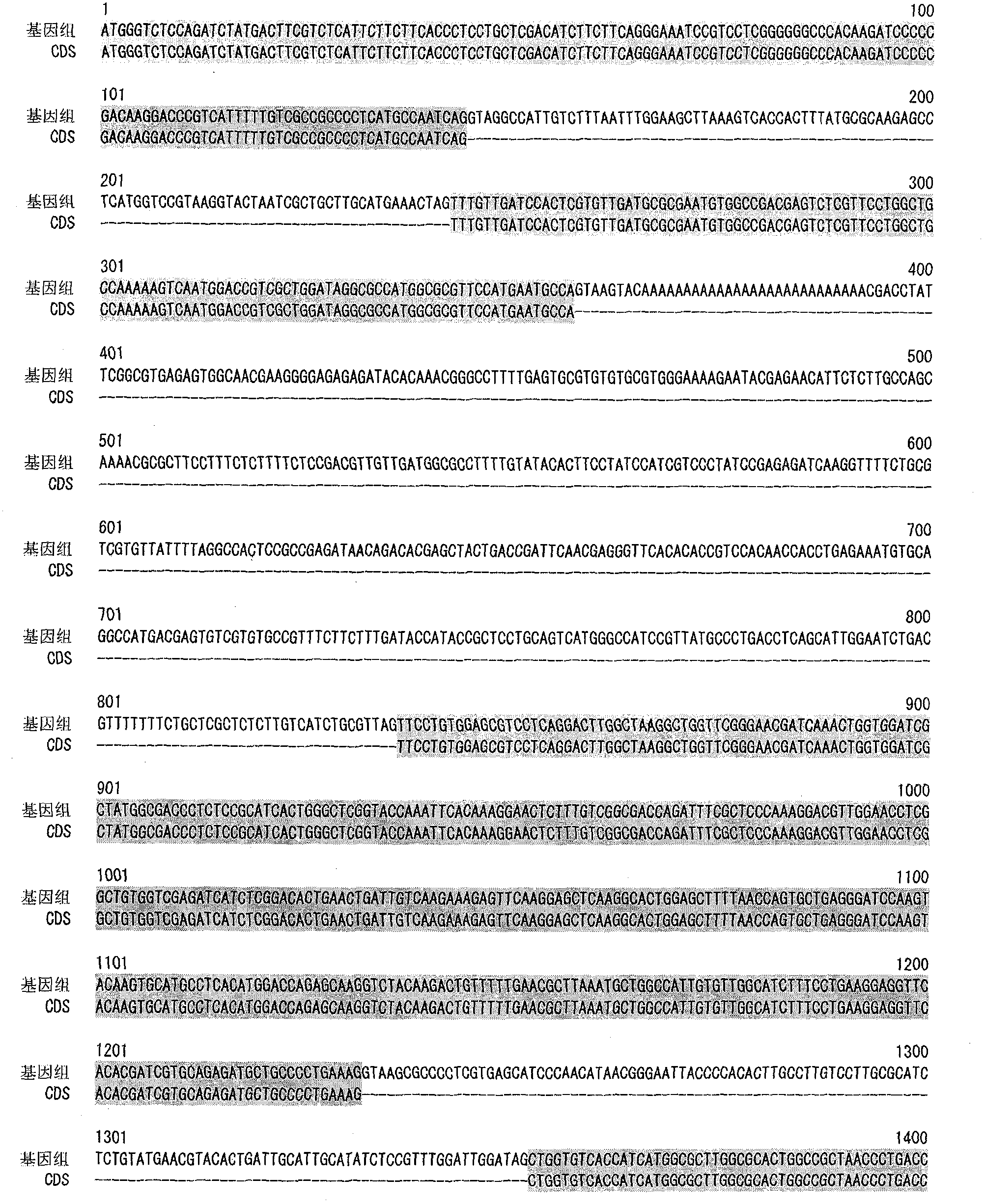
[0120] Genome Analysis of M.alpina
[0121] M.alpina 1S-4 strain was inoculated into 100 ml of GY2:1 medium (2% glucose, 1% yeast extract, pH 6.0), and cultured with shaking at 28°C for 2 days. The bacterial cells were collected by filtration, and genomic DNA was prepared using DNeasy (QIAGEN).
[0122] The base sequence of the above genomic DNA was determined using Roche 454 Genome Sequencer FLX Standard. In this case, the determination of the base sequence of the fragment library was performed twice (run), and the determination of the base sequence of the paired-end library was performed three times of sequencing (run). By splicing the obtained base sequences, 300 super contigs (Super Contig) were obtained.
[0123] Synthesis of cDNA of M.alpina 1S-4 Strain
[0124] M.alpina 1S-4 strain was inoculated into 4 ml of medium (2% glucose, 1% yeast extract, pH 6.0), and cultured at 28° C. for 4 days. Bacteria were recovered by filtration, and RNA was extracted using RNeasy...
Embodiment 2
[0136] Complementation experiment of yeast S.cerevisiae (Δsct1, Δgpt2)
[0137] In the yeast S. cerevisiae, SCT1 and GPT2 are known as genes responsible for GPAT activity, and simultaneous deletion of these genes leads to lethality. In order to confirm whether the proteins encoded by MaGPAT1 and MaGPAT3 from M.alpina have GPAT activity, the complementation experiments of Δsct1 and Δgpt2 were carried out. Table 1 shows the genotypes of the strains produced in this experiment.
[0138] [Table 1]
[0139]
[0140] Production of GP-1 strain
[0141] The SCT1 gene of Δgpt2 homozygous diploid yeast (product number YSC1021-663938) of a yeast knockout strain (product name: yeast knock out strain collection) (Open Biosystems) was disrupted by the following method. First, DNA was extracted from the cells of S. cerevisiae S288C strain using Dr. GenTLE (Gen とる く ん) (for yeast) (TAKARA BIO). Using this as a template, using primer Xba1-Des-SCT1-F: 5'-TCTAGAATGCCTGCACCAAAACTCAC-3 (...
Embodiment 3
[0160] Fatty acid analysis of yeast
[0161] The strains GP-12 (#1, 2, 3), MaGPAT1-11 (#3b, #4a, #8a), MaGPAT3-11 (#2d, #19a, #32a), GP-22 obtained by the above operation (#1, #2, #3), MaGPAT1-21 (#1a, #2d, #13a), MaGPAT3-21 (#10a, #20c, #26b) were cultured as follows.
[0162] 10 ml of SD-Ura and Leu liquid culture medium were inoculated with each bacterial strain of 1 platinum fungus, and cultured with shaking at 30° C. for 1 day. 100 μl of the obtained culture solution was inoculated into 10 ml of SD-Ura and Leu liquid medium, and cultured with shaking at 30° C. for 2 days. The bacterial cells were recovered by centrifuging the yeast culture solution. The bacterial cells were washed with 10 ml of sterilized water, centrifuged again to recover the bacterial cells, and freeze-dried. After the fatty acid in the bacterial cell was converted into methyl ester by the hydrochloric acid methanol method, it was extracted with hexane, the hexane was distilled off, and the analys...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com